Max Planck Institute for Solid State Research, Heisenbergstrasse 1, D70569 Stuttgart, Germany. b.kannan@fkf.mpg.de
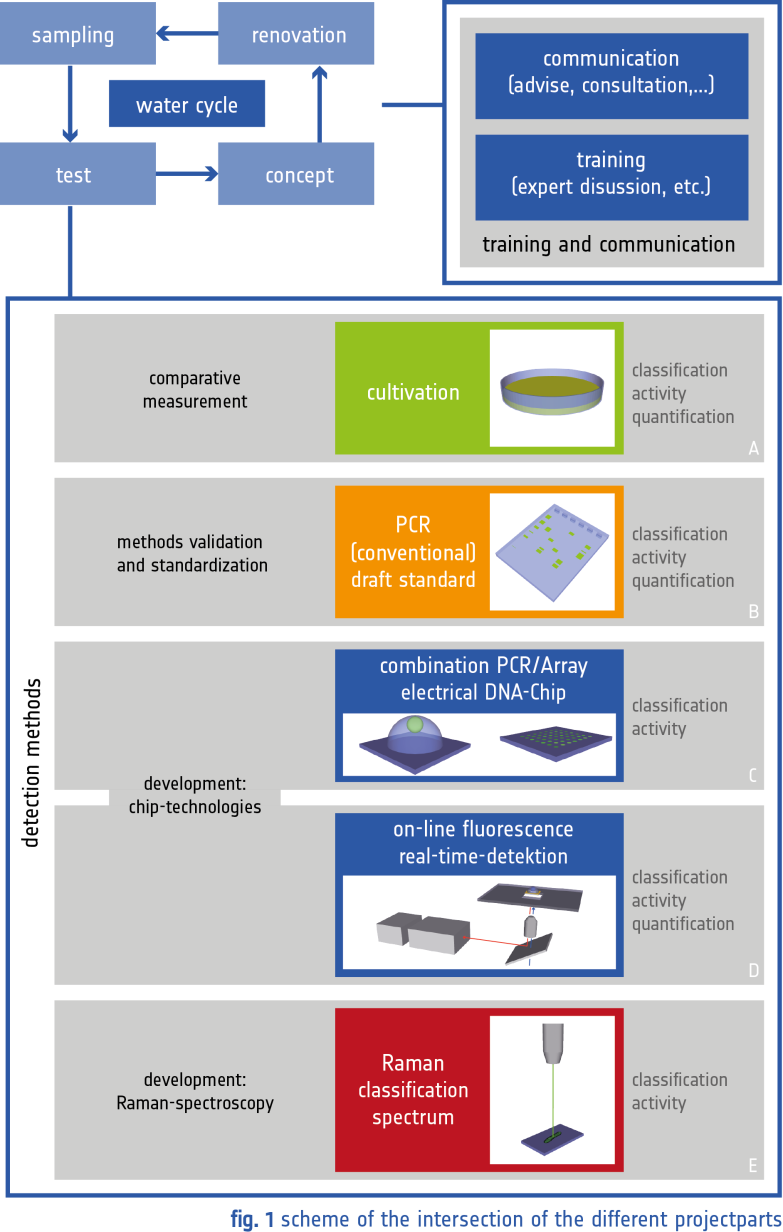
The specific detection of nucleic acids down to a few molecules is a central cornerstone of modern bioanalytical science. Many of the currently prevalent methods are based on the optical detection of labelled target sequences, utilizing a pre-amplification step based on polymerase chain reaction (PCR). Label-free methods such as sandwich assays are also widely used, however they require many processing steps and rely on more than one binding event. [1] In order to obtain label-free sensors with a small footprint and to attain fast response times, direct one-step electrical detection methods are being experimented widely. Nanostructures such as carbon nanotubes (CNTs) are ideal candidates for this purpose, since they offer a very high surface-to-volume ratio thereby showing promise for the realization of sensitive biodetectors. They are unique in the sense that few binding events occurring on their nanoscale surface can be directly transduced to an electrical signal. [2] A key challenge in the realization of such nanobiosensors is the possibility to attain selectivity while maintaining the high sensitivity of such devices. The talk will present a strategy that we have adopted in overcoming this challenge. Specifically CNT device arrays are fabricated by scalable lithographic techniques. In order to attain selectivity the probe DNA sequence is attached using a specialized electrochemical functionalization strategy. [3] This ensures that the probe DNA is exclusively coupled to the CNT surface (Figure 1A). The detection is based on a unique impedance measurement technique coupled to electrical field effect. This provides us with a good degree of stability and reproducibility of the DNA nanosensors. We demonstrate that using this strategy we can directly detect DNA down to attomolar concentrations (Figure 1B) without the use of markers or sandwich protocols. [3] Calibration and reproducibility issues, and the ability to detect DNA in a complex matrix such as serum will also be discussed.
- Fig. 1: A Schematic of the CNT-based DNA nanosensor – a CNT contacted by two electrodes is functionalized with a probe DNA sequence using an intermediate polymer layer. The target sequences bind specifically on to the CNT surface inducing a sensor response. B Sensor signal (threshold voltage) as a function of DNA concentration for complementary target (cDNA) and for a non-complementary sequence ( ncDNA).
[1] Kurkina, T. & Balasubramanian, K. 2012 Towards in vitro molecular diagnostics using nanostructures. Cell. Mol. Life Sci. 69, 373.
[2] Vlandas, A., Kurkina, T., Ahmad, A., Kern, K. & Balasubramanian, K. 2010 Enzyme-free sugar sensing in microfluidic channels with an affinity-based carbon nanotube sensor. Anal. Chem. 82, 6090
[3] Kurkina, T., Vlandas, A., Ahmad, A., Kern, K. & Balasubramanian, K. 2011 Label-free detection of few copies of DNA using carbon nanotube impedance biosensors. Angew. Chem. Intl. Ed. 50, 3710.
Centro di Riferimento Oncologico (CRO), Experimental anc Clinical Pharmacology Unit, Via Franco Gallini 2, 33081, Aviano, Pordenone, Italy and Temple University, Department of Biology, 1900 N. 12th street, 19122 Philadelphia, PA, USA mcastronovo@cro.it
Nanotechnology allows to confine DNA molecules into packed systems where the level of crowding is closer to the one in cells. The effect of nanometric confinement on the transport and the biochemical properties of biomolecules within these nanostructures is however substantially unknown.
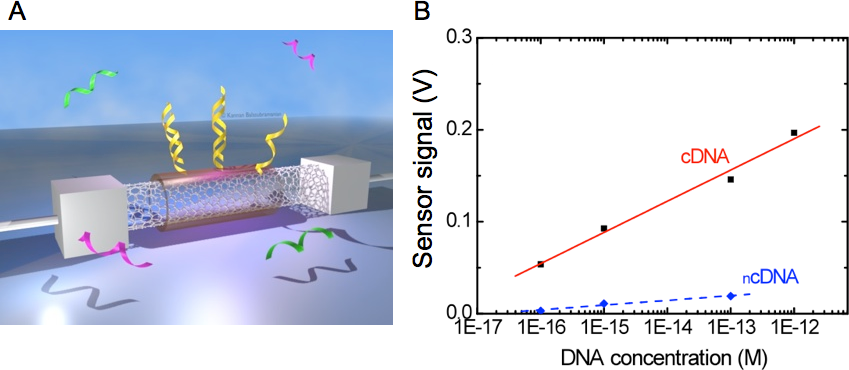
In our experimental work we studied the mechanism by which restriction enzymes work inside brushes of short double stranded (ds)DNA molecules, confined on ultra-flat gold surfaces by using nanografting, an atomic force microscopy (AFM) based nanolithography method (see Figure 1.A-D). dsDNA molecules have a restriction site at half height and, in turn, successful restriction reactions lead to a 50% decrease of the brush height with respect to the surrounding surface, that we measure by AFM (Fig.1 F). We address the effect of confinement on these reactions by varying the dsDNA density, and we unequivocally show that the confinement has a quantifiable effect on the mechanisms of enzyme diffusion and reaction.
1) We show that enzyme molecules do not access to the dsDNA brush directly from the solution, but two dimensionally diffuse inside the brush, and exclusively access from the sides (see Fig.1 G). Moreover, the access is arrested when the DNA density reaches a certain critical threshold.
2) We show that when the dsDNA density is sufficiently high, the restriction reaction can successfully occur on a dsDNA site having only partial consensus for the enzyme.
Our findings demonstrate that, in crowded systems, enzyme may work very differently than in solutions. We demonstrate the possibility to control the diffusion of enzymes, inside surface-bound DNA nanostructures. In general our findings are opening the door to novel applications of DNA nano-assemblies in both the fields of biosensing and fundamental biophysics.
- Fig. 1: A-C Schematic view of the DNA nanografting method. B – One surface bound brush of ssDNA is generated. D – The DNA brush is hybridized. The dsDNAs have a specific restriction site in the middle (in orange). The absolute initial height (IH) of the dsDNA matrix is determined by AFM imaging at low forces (blue arrow). E – An enzymatic restriction reaction cleaves the DNAs within the brush. F – The absolute final height of dsDNA brush is measured by AFM. G – Two-dimensional enzyme diffusion within DNA nanostructures. On the top, enzymes cannot access the low-density brush due to the high-density structure that surrounds the core. On the bottom, the low density brush is accessed by the enzyme molecules which cleave the inherent DNAs.
[1] Castronovo, M., Lucesoli, A., Parisse, P., Kurnikova, A., Malhotra, A., Grassi, M., Grassi, G., Scaggiante, B., Casalis, L., Scoles, G., Two-dimensional enzyme diffusion in laterally confined DNA monolayers. Nature Communications 2011, 2, 297.
Institute of Photonic Technology, Albert-Einstein-Straße 9, 07745 Jena, Germany
Inst. Physical Chemistry, University of Jena, Helmholtzweg 4, 07743 Jena, Germany
volker.deckert@uni-jena.de
Plasmonic particles at the apex of AFM probes are used to amplify electromagnetic fields at the tip when the particles are irradiated with laser light.[1,2] The field enhancement leads to a further enhancement of Raman signals of the sample by several orders of magnitude. The enhancement can be high enough to enable even single molecule detection. A further property of such an experimental arrangement is the local confinement of the involved electromagnetic fields. Only a small volume, related to the diameter of a plasmonic particle is excited resulting in a lateral resolution exceeding all other optical methods used at present.
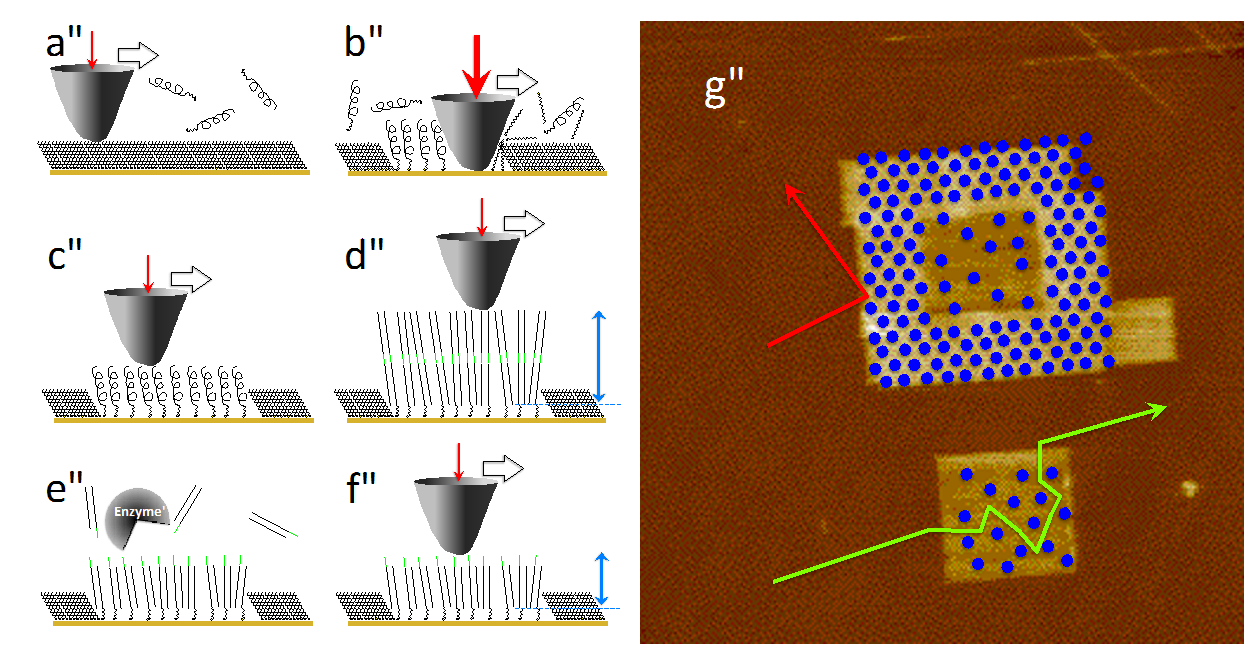
We will demonstrate the capabilities of this so-called tip-enhanced Raman scattering (TERS) when applied to chain-like bio molecules like DNA or peptides.[3-6]}. Specifically the potential of sub-nanometer resolution will be demonstrated (see Fig 1) and the potential of a direct sequencing of DNA will be addressed and potential pitfalls will be discussed.
- Fig. 1: Intensity distribution of specific amino acids on an insulin fibril along a TERS line scan. Distance between positions 0.5 nm, respectively. The size of the spots corresponds to the detected normalized intensity of each band.
[1] Stöckle, R., Suh, Y., Deckert, V. & Zenobi, R. Nanoscale chemical analysis by tip-enhanced Raman spectroscopy. Chem Phys Lett 318, 131–136 (2000).
[2] Bailo, E. & Deckert, V. Tip-enhanced Raman scattering. Chem. Soc. Rev. 37, 921–930 (2008).
[3] Bailo, E. & Deckert, V. Tip-enhanced Raman spectroscopy of single RNA strands: towards a novel direct-sequencing method. Angew Chem Int Ed Engl [ 1658–1661 (2008).
[4] Treffer, R. & Deckert, V. Recent advances in single-molecule sequencing. Curr. Opin. Biotechnol. 21, 4–11 (2010).
[5] Treffer, R., Lin, X., Bailo, E., Deckert-Gaudig, T. & Deckert, V. Distinction of nucleobases - a tip-enhanced Raman approach. Beilstein J Nanotechnol 2, 628–637 (2011).
[6] Deckert-Gaudig, T., Kämmer, E. & Deckert, V. Tracking of nanoscale structural variations on a single amyloid fibril with tip-enhanced Raman scattering. J. Biophoton. 5, 215–219 (2012).
aCavendish Laboratory, University of Cambridge, JJ Thomson Ave, Cambridge, CB3 0HE United Kingdom
bCenter for Nanoscience and Department of Physics, Ludwig-Maximilians-Universität München, Geschwister-Scholl-Platz 1, 80539 München, Germany
cDepartment of Materials Science and Metallurgy, University of Cambridge, Pembroke Street, CB3 3QZ Cambridge, United Kingdom
Nanopores in insulating membranes provide the possibility to detect single molecules in solution[1]. Common techniques in solid-state nanopore production, like transmission-electron drilling, however do not give the opportunity to control the exact shape of the cavity.
Encouraged by Hall et al.[2], demonstrating the combination of a protein and solid-state nanopore, we show a new way for custom-tailored cavities built up entirely from DNA using the DNA origami method. This well established method allows creating almost arbitrary structures on the nanometer scale by utilizing the programmability of DNA self-assembly. Here a 7-8kb long m13mp18-based single strand of DNA is formed by hundreds of short synthesised DNA strands, so-called staple strands, into a desired shape.
Our DNA origami nanopore is a conical structure composed of four overlapping skirts with a quadratic base (Figure 1A). The chosen design is intended to fit the conical form of the solid-state nanopore with the goal to keep the leakage at the interface and through the DNA structure itself minimal. Furthermore we added a 2344bp long DNA overhang to guide the voltage driven hybrid pore formation (Figure 1B & 1D). Due to the easy modifiability of the shape of DNA origami structures we were able to incorporate a flap into the design.
We will present detailed aspects of the DNA origami design as well as experimental data showing the successful and stable insertion of a DNA origami nanopore into a solid-state pore (Figure 1C & 1D). Furthermore we will show controllably and repeatedly hybrid pore formations. Finally we demonstrate the detection of -DNA with our hybrid pores[3].
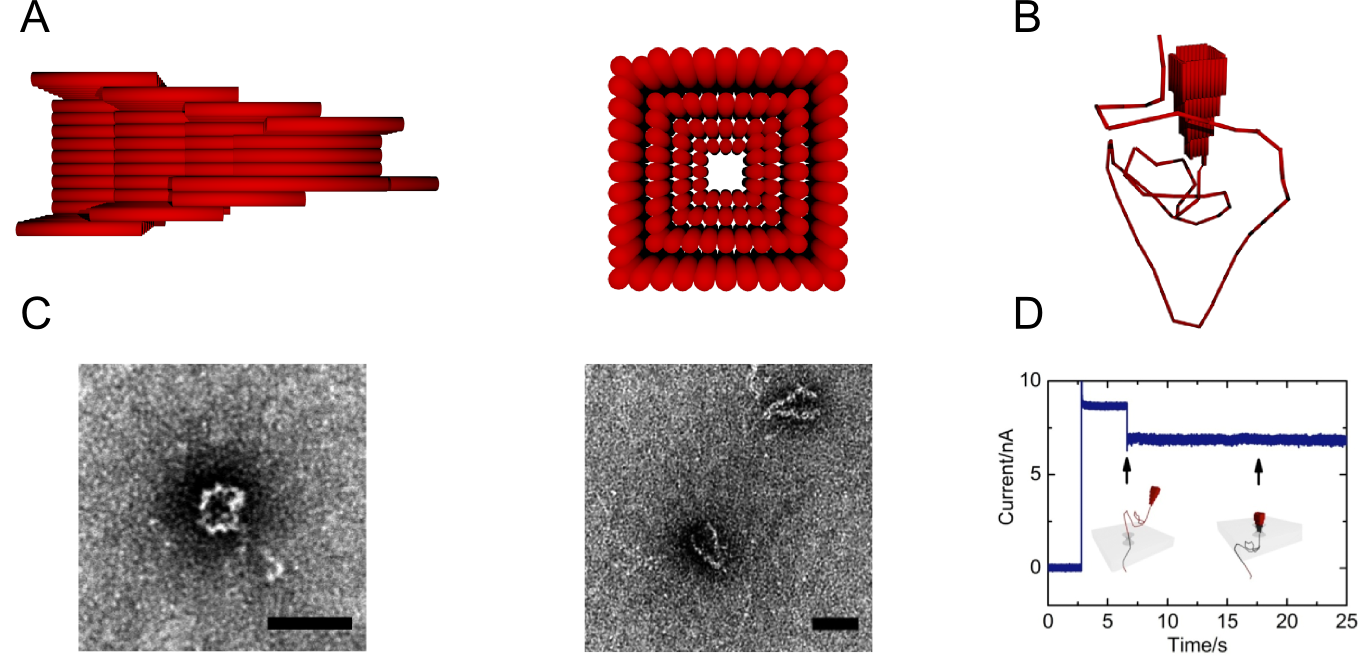
- Fig. 1: A - Schematic cross-section and top view of the DNA origami nanopore.
B – Schematic DNA origami nanopore design illustrating the DNA overhang. C - Top view (left) and side view (right) TEM micrographs of the DNA origami nanopore, scale bar = 50 nm. D – Experimental Data indicating a hybrid pore formation.
[1] Kasianowicz et al., Proc. Natl. Acad. Sci. U.S.A. 1996, 93 (24), 13770-13773.
[2] Hall et al., Nat. Nanotechnol. 2010, 5 (12), 874-877.
[3] Bell, N. A. W., Engst, C.R., Ablay. M., Divitini, G., Ducati, C., Liedl, T., Keyser, U.F. 2012 DNA Origami Nanopores. Nano Lett. 2012, 12(1), 512-517.
Interdisciplinary Nanoscience Center and Center for DNA Nanotechnology, Aarhus University, Ny Munkegade 1521, DK-8000 Aarhus C, Denmark elena.ferapontova@inano.au.dk
Genosensor technologies have become increasingly important in molecular diagnostics, both for diagnosis and preventative therapy of genetic disorders, prognosis of cancer, and treatment of infections. In this context, combination of electrochemistry and DNA nanotechnology allowed the development of extremely sensitive and accurate, yet simple, inexpensive and robust genosensor platforms [1,2]. In particular, redox-labeled DNA beacon systems allow developing practically attractive label-free electrochemical genosensors. In such genosensors, hybridization of target DNA to the conformationally robust probe DNA sequence tethered to the electrode surface results in the electrochemically addressable conformational nanoswitching of the probe [3-5]. Therewith, an electrochemical signal stemming from the reaction between the redox label and the electrode is generally considered as the evidence of the electronic conductivity of dsDNA, and genosensors are designed based on that concept. However, in cases when intercalation of redox probes into the DNA duplex does not occur, data on directional electron transfer (ET) between electrodes and DNA-conjugated redox probes are inconsistent [5,6] . In this paper it is discussed how ET in the electronic DNA hairpin molecular beacons can be modulated for the development of simple and robust genosensor platforms with improved selectivity for single nucleotide polymorphism (SNP). We show that in loosely packed DNA monolayers ET between the electrodes and the redox probes conjugated to dsDNA is governed by the nanoscale motion of the probe (and thus DNA) towards the electrode surface (Figure 1). Therewith, the mode of the redox probe conjugation to DNA and its charge affects kinetics of ET [7]. Complex mechanism of ET between the electrode and differently charged redox probes conjugated to DNA by different means will be discussed, and applications for analysis of a cancer biomarker TP53 gene will be shown.
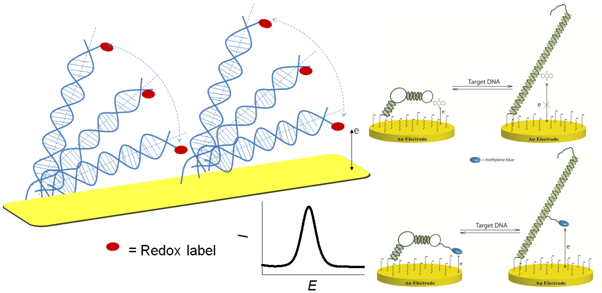
- Fig. 1: Nanoscale movements of the DNA duplex are responsible for the redox signal correlating with a diffusional mechanism of electron transfer. Those movements depend on the charge and redox potential of the redox probe used for DNA labeling, as well as on the way the redox probe is linked to DNA: for the uncharged anthraquinone probe no electrochemical signal can be detected in the DNA hybrid, while for positively charge methylene blue there is a strong ET signal due to diffusional movements of the probe towards the negatively charged electrode surface.
[1] Drummond T.G., Hill M.G., and Barton J.K., 2003 Electrochemical DNA sensors, Nature Biotech. 21, 1192.
[2] Ferapontova E.E., 2011 Electrochemical indicators for DNA electroanalysis, Curr. Anal. Chem. 7, 51.
[3] Fan C., Plaxco, K.W., and Heeger, A.J., 2003 Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA, Proc. Natl. Acad. Sci. USA 100, 9134.
[4] Farjami E., Clima L., Gothelf K.V., and Ferapontova E.E., 2010 DNA interactions with a methylene blue redox indicator depend on the DNA length and are sequence specific, Analyst 135, 1443.
[5] Farjami E., Clima L., Gothelf K.V., 2011 An "off-on" electrochemical hairpin DNA-based genosensor for cancer diagnostics, Anal. Chem. 83, 1594.
[6] Anne, A. and Demaille, C., 2006 Dynamics of electron transport by elastic bending of short DNA duplexes., J. Am. Chem. Soc. 128, 542
[7] Abi A. and Ferapontova E.E., Unmediated by DNA electron transfer in redox-labeled DNA duplexes end-tethered to gold electrodes,
University of California San Diego, Departments of Bioengineering and Nanoengineering, Atkinson Hall Rm 2312, La Jolla, CA, 92093-0448, mheller@ucsd.edu, 760-415-1962 (c)
While an on-line search for "DNA Biosensors" yields more than 40,000 hits, the actual number of DNA sensor devices that are used for clinical diagnostics are minimal. Two basic reasons for this poor success rate for technology translation are sample preparation and inaccuracy of base discrimination. These two problems limit the success of DNA sensor devices far more often than does intrinsic detection sensitivity. Apparently, these problems may also be limiting new DNA sequencing technologies, including single molecule nanopore sequencing, for which $billions have been spent. We now show that a microarray dielectrophoretic (DEP) device can be used to rapidly isolate and detect disease related DNA biomarkers, bacteria, virus and nanoparticles directly from undiluted whole blood. At DEP frequencies of 5kHz-10kHz both hmw-DNA and any nanoparticulate (20nm-700nm) biomarkers are separated from the blood and become highly concentrated at specific DEP high field regions over the microelectrodes, while blood cells move to the DEP low field regions. The blood cells can then be removed by a simple fluidic wash while the hmw-DNA and nanoparticles remain highly concentrated in discrete microscopic locations. At this point, in-situ PCR or other detection procedures (fluorescent dyes, hybridization, etc,) can be carried out in the same device chamber. Presently, these sample to answer devices are being used for the rapid detection of cancer related mutations in cell free circulating (cfc) DNA isolated directly from the blood of patients with Chronic Lymphocytic Leukemia (CLL) and brain tumors; as well for the complete sample preparation of bacterial DNA for DNA sequencing applications.
Michael J. Heller received his Ph.D. in Biochemistry from Colorado State University in 1973. He was an NIH Postdoctoral Fellow at Northwestern University from 1973 to 1976. Dr. Heller was supervisor of the DNA Technology Group at Amoco Corporation from 1976 to 1984. During that time he carried out bioengineering and recombinant DNA engineering work on plants, algae and photosynthetic bacteria for energy and chemical production, and oversaw the companies sponsored research at the Cetus Corporation. Dr Heller was Director of Molecular Biology at Molecular Biosystems from 1984 to 1987. He was a co-founder of Integrated DNA Technologies and served as President and Chief Operating Officer from 1987 to 1989. Dr Heller was a co-found of Nanotronics and Nanogen and served as the Chief Technical Officer from 1993 to 2001. Nanogen carried out the successful development of microelectronic DNA array technology for clinical genotyping applications. Dr. Heller is now a Professor in the Departments of Nanoengineering and Bioengineering at the University California San Diego. He has also recently co-founded a new company called Biological Dynamics (started by his PhD student) which is developing new cancer diagnostics technology. Dr. Heller has extensive industrial experience in biotechnology, biomedical and clinical diagnostic devices and nanotechnology; with particular expertise in the areas of DNA probe diagnostics, DNA synthesis, fluorescent-based detection technologies and electric field assisted self-assembly of DNA nanocomponents. Dr. Heller has a respectable publication record, has over 45 issued US Patents and has been an invited speaker to a large number of scientific conferences and meeting. Dr. Heller has been a panel member for the White House (OSTP) National Nanotechnology Initiative 1999/2000; the NAS (NAE) Review of National Nanotechnology Initiative 2001-2002; the NAS(NAE) – Engineer for the 2020 - 2001/2002; and has also been involved in a number of NSF Nanotechnology Workshops.
Service des Photons, Atomes et Molécules, Laboratoire Francis Perrin (CEA-CNRS URA 2453)
CEA Saclay, 91191 Gif/Yvettte Cedex, France
Nanomaterials have applications in various fields such as microelectronics, electromagnetism, optoelectronic and chemistry (biomedical, pharmaceutical, cosmetics...) and also in the field of environmental catalysis, i.e. new processes for green chemistry. Catalysis (or photocatalysis) is one of the privileged applications in nanoscience. Indeed, the high dispersion of the active phase (oxide, metal) strongly increases the catalytic efficiency as well as the durability of the solids.
In order to get such nanomaterials, laser pyrolysis is a promising and versatile method allowing the synthesis of various nanoparticles, with well defined chemical composition, size and structure [1]. This gas phase synthesis method is based on the interaction of a powerful IR laser beam with a mixture of gaseous or liquid precursors, one of them absorbing the laser radiation. The temperature increases quickly, leading to the decomposition of the precursors. An incandescent flame is observed in the interaction zone where nanoparticles nucleate and grow. The residence time adjusted by the carrier gas flow allows the control of the nanoparticle size. The chemical composition depends on the nature and the content of the different reagents in the precursor mixture.
To synthesize the Titanium oxide based materials, TTIP (Titanium Tetra isopropoxide) was chosen as Ti precursor. It was possible to adjust the TiO2 anatase to rutile ratio '10/90 to 90/10) by adjusting the reaction temperature through a control of experimental parameters such as laser power [2]. It was also possible to produce M@TiO2 nanoparticles with M=Pt, Pd, Fe, Ag. To achieve such synthesis, a solution containing the precursors was prepared by first dissolving appropriate amounts of metal acetylacetonate (for example Pt(Acac) or Pd(Acac)) in a xylene/ethylacetate mixture (65:35 vol/vol) and then mixing it with TTIP. Nevertheless such mixture absorbs only weakly the laser radiation so ethylene C2H4 was added as sensitizer gas and the mixture was sent to the reaction zone as an aerosol. We also compared the catalytic activity of nanoPd/TiO2 and nanoPt/TiO2 [3]. For photocatalytic applications, many papers concerns the development of materials exhibiting an optical absorption in the visible range and N-TiO2 nanoparticles have achieved special interest. Using the laser synthesis method, such nanoparticles could easily be obtained just by adding a flow of NH3 in the reactant mixture [2]. Moreover, by increasing the NH3 flow, it was possible to obtain highly reducing conditions in the reaction zone and crystallized TiOx (X=1 to 2) nanoparticles were synthesized [4], such nanoparticles exhibit a large shift (up to 2 eV) of their absorption in the visible range. Their interest for photocatalysis is under study.
[1] W.R. Cannon, S.C. Danforth, J.S. Haggerty and R.A. Marra, J. Am. Ceram. Soc, 65, 330 (1986)
[2] B. Pignon, H. Maskrot, Y. Leconte, C. Reynaud, N. Herlin-Boime, V. Guyot Ferreol, T. Pouget, JF Tranchant M. Gervais S. Coste, European Journal of Inorganic chemistry, 208 (6), 883-889, 2008
[3] H. Maskrot, Y. Leconte, N. Herlin-Boime, C. Reynaud, E.Guelou, L.Pinard, S.Valange, J. Barrault, M. Gervais, Catalysis Today, 2006, 116 (1): 6-11. doi:10.1016/j.cattod.2006.04.006
[4] P. Simon, B. Pignon, B. Miao, S. Coste, Y. Leconte, S. Marguet, P. Jegou, B. Bouchet-Fabre, N. Herlin-Boime, N-doped titanium monoxide nanoparticles with TiO rock-salt structure, low energy band gap and visible light activity" Chem mater 2010, 22(12), 3704-3711, (2010)
Chemical Nanoscience Laboratory, School of Chemistry, Bedson Building, Newcastle University, Newcastle Upon Tyne, NE1 7RU, UK
Recently nucleobases have been used to produce 1D coordination polymers which form structures similar to that of DNA. In this poster a new route to modify oligomers is reported. The on-column thio-conversion of guanine and the synthesis of modified oligomers were studied to investigate metal binding in order to explore the formation of 1D coordination polymers, summarised below in Fig. 1.
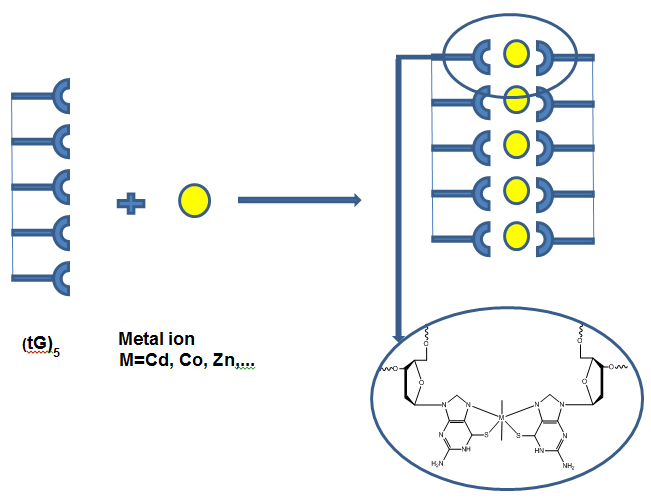
- Fig. 1: Synthetic route for the synthesis of thio-functionalized oligomers for metal binding coordination polymers.
Nanoscience Center, 1Department of Physics and 2Department of Chemistry, P.O.Box 35 (YN) 40014 University of Jyväskylä, Finland
3Department of Applied Physics, Aalto University School of Science, P.O.Box 15100, 00076 AALTO, Finland tommi.isoniemi@jyu.fi
Field-effect transistors (FETs) with carbon nanotubes (CNTs) as their conductance channel have possible applications as optical and chemical sensors. Chemical sensing is generally based on adsorption and desorption of different molecules that alter the electrical response of semiconducting CNTs, and several photoinduced effects can also be used for gating. Surface plasmon polaritons (SPPs) are coupled modes of electromagnetic waves and collective electron excitations at a metal surface, and they can be considered as two-dimensional light bound to the interface. They offer fascinating prospects in photoelectronics due to their high confinement of optical fields.
In this work we demonstrate a novel CNT FET that can be modulated with SPPs, utilizing changes in the chemical environment to control the charge transfer properties [1]. SPP excitation is done via the Kretschmann configuration while the measured CNT FET is situated within reach (50 nm) of the SPPs. Using a 633 nm laser producing a SPP power density of 110 mW/cm2 we observe a shift of ≈0.4 V in effective gate voltage. SPP-intermediated desorption of physisorbed oxygen from the device is a likely mechanism for the effect, and field enhancement at the metal surface results in a lower required excitation intensity compared to far-field photodesorption.
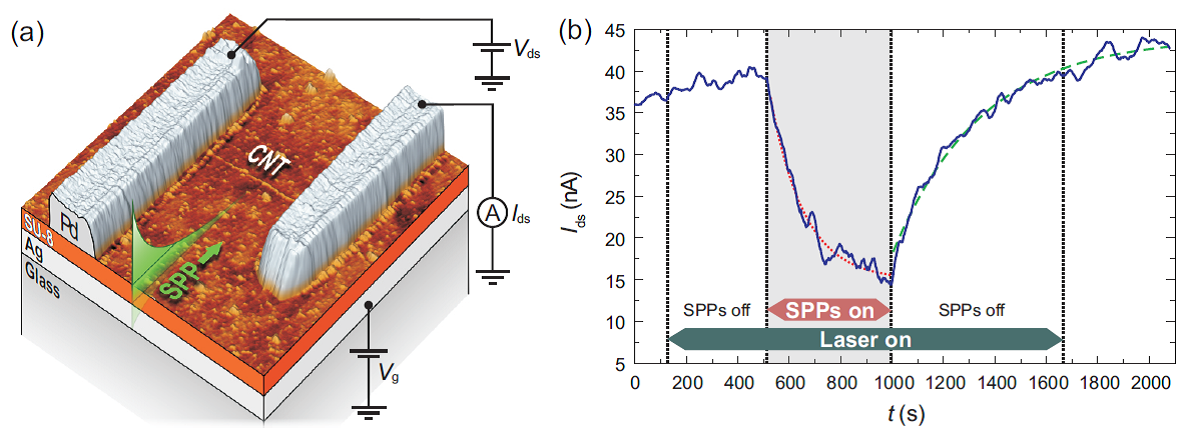
- Fig. 1: (a) - AFM image and schematic of the device. SPPs are launched on the Ag-SU-8 interface at the exposed 1 µm long CNT by laser excitation through the glass. (b) - Current response to laser exposure and SPP excitation at 1.96 eV.
[1] Isoniemi, T., Johansson, A., Hakala, T.K., Rinkiö, M., Törmä, P., Toppari, J.J. & Kunttu, H. 2011 Surface plasmon effects on carbon nanotube field effect transistors. Applied Physics Letters 99, 031105
Institute of Photonic Technology, Albert-Einstein-Straße 9, 07745 Jena, Germany
jacqueline.jatschka@ipht-jena.de
In the field of bioanalytics, noble metal nanoparticles offer a fast and reliable recognition of biomolecules due to their unique optical properties. Their distinctive feature is caused by density oscillations of their conduction electrons due to external electromagnetic irradiation, also called Localized Surface Plasmon Resonance (LSPR). This occurring resonance band can be adjusted by shape, size and material of the nanoparticle and influenced by changes of the local refractive index of the surrounding medium. This fact gives the opportunity to use noble metal nanoparticles as label-free bioanalytical sensors. Biomolecules can be bound directly on the nanoparticle's surface, which leads to a change of the local refractive index and a shift of the maximum peak detected by micro-spectroscopy. (Figure 1)
One interesting aspect is the analysis of DNA-DNA interaction on LSPR-sensors. On one hand the attachment of the capture DNA strands and on the other hand the sequence specific DNA binding (hybridization) under different conditions were analyzed. For instance the often used DNA sandwich system was applied on a single nanoparticle level. Additionally the detection limit of the single stranded DNA on the LSPR sensor system was determined.
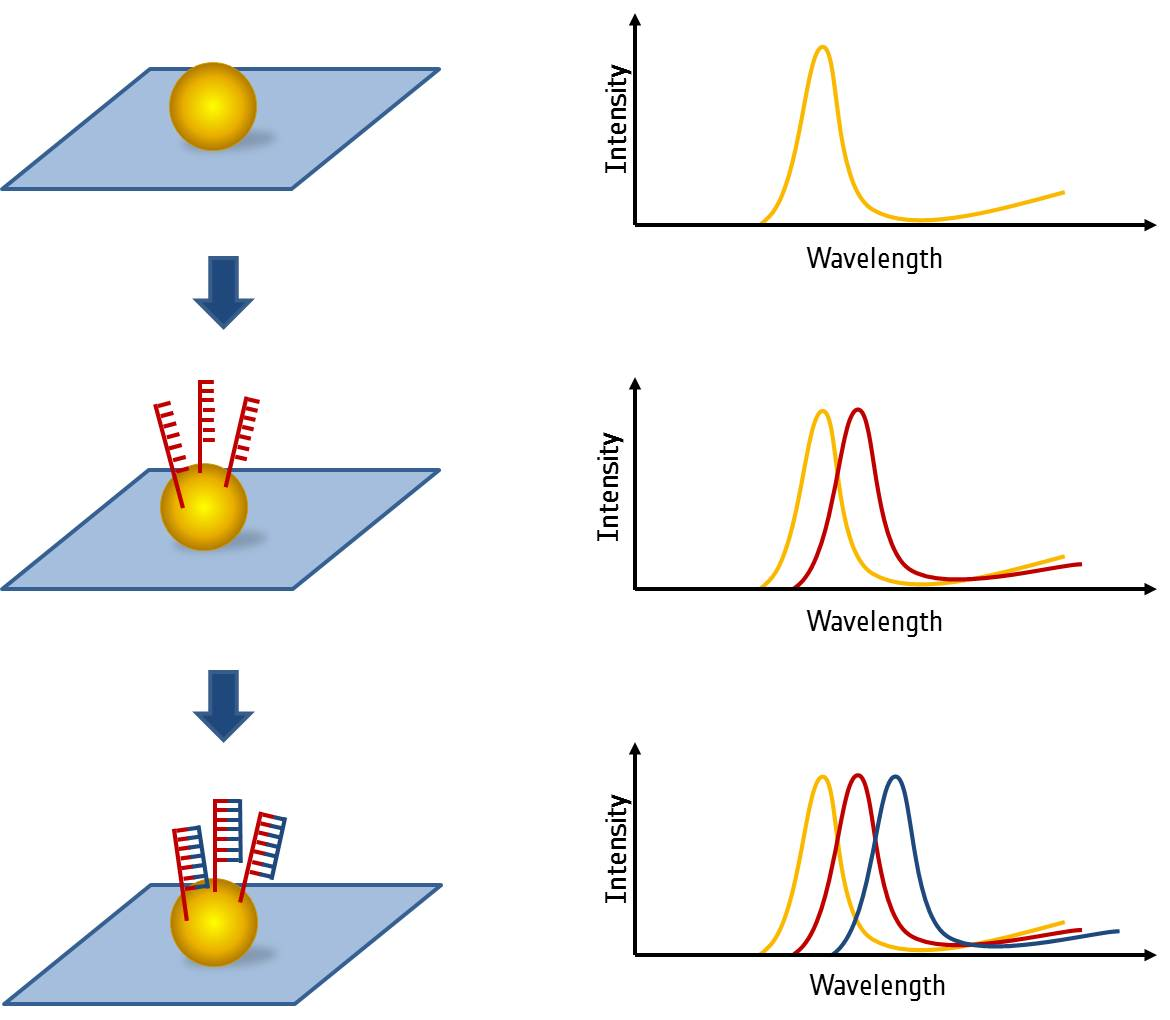
- Fig. 1: Schematics of the LSPR sensing principle on single nanoparticle level
[1] Hutter, E. and J. H. Fendler (2004). Exploitation of Localized Surface Plasmon Resonance. Advanced Materials 16(19): 1685-1706.
[2] Schneider T., et al. (2010). Nanoparticles and DNA on Chips – Realization of a Nanobiotechnology Toolbox for single particle bioanalytics. Leibniz-Institut LIFIS Online. ISSN 1864-6972.
Institute of Photonic Technology, Albert-Einstein-Straße 9, 07745 Jena, Germany
DNA-Origami has become to one of the most noted methods in DNA-Nanotechnology. With this bottom up technic nanoscale DNA shapes can be easily formed by self-assembly [1]. In this process one long single stranded DNA molecule gets folded by many different short staple stands. DNA-Origamis can also connect to each other specifically [2]. By modifing single staple strands the DNA Origami can serve as a nano-breadboard for e.g. metal nanoparticles with a spatial resolution of roughly 6 nm.
Metal-nanoparticle structures are interesting for their optical properties caused by plasmonic effects. Incident electromagnetic waves induce a collective oscillation of the conduction electrons in subwavelength particles. This excitation can interact with other particles in very short distances by plasmonic resonance effects. So in theory an accurately defined nanoparticle pattern could function as a "plasmonic fiber" or as an amplifier [3]. Therefore the combination of DNA-Origami structures with their features of nanoscaled definition options and metal nanoparticles represents a promising next logical step.
We also present our experiments for the defined metallization of 2D-DNA Origami structures using gold-ions in solution. With an easy and reliable method for DNA metallization, DNA and its superstructures could serve as a template for nanoscale electrical and photonic devises.
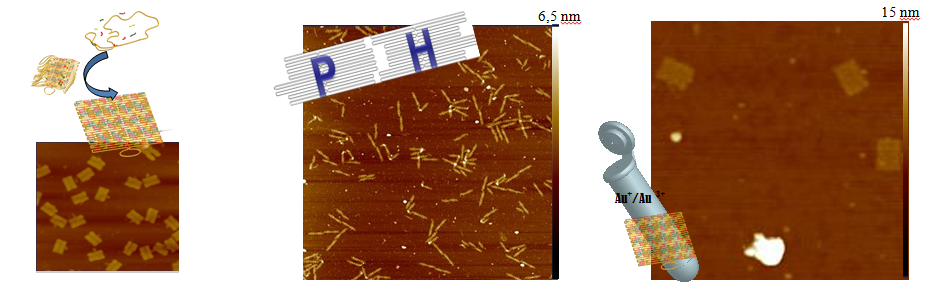
- Fig. 1: A: Prinziple of DNA-Origami self-assembly process. B: specific combination of two DNA-Origami labelled with DNA-harpins ([2] modified, scan-size 5 µm) C: Mixture of normal DNA-Origami rectangles and rectangles treated with gold-ions solution (scan-size: 850 nm).
[1] P. Rothemund (2006). "Folding DNA to create nanoscale shapes and patterns". Nature 440 (7082): 297–302.
[2] S.Woo (2011). "Programmable molecular recognition based on the geometry of DNA nanostructures" Nature Chemistry
(VOL 3) :620-627
[3] S.Bidault, F.J. Garcia de Abajo, A. Polman, Plasmon-Based Nanolenses Assembled on a Well-Defined DNA Template, J. Am. Chem. Soc.,2008
Laboratory of Bioanalytical Chemistry, Department of Chemistry, Adam Mickiewicz University, Grunwaldzka 6, 60-780 Poznan, Poland, juskowia@amu.edu.pl
*Institute of Photonic Technology, Albert-Einstein-Straße 9, 07745 Jena, Germany
DNAzymes with peroxidase-mimicking activity found wide application in biosensing. Due to their advantages in comparison with protein enzyme (thermal stability, simpler obtaining and purification) they can successfully replace horseradish peroxidase in bioanalytical applications [1]. DNAzymes with peroxidase activity must adopt G-quadruplex topology and form complex with hemin in order to perform catalytic reaction. Such a system can successfully catalyze redox reaction between hydrogen peroxide and a suitable substrate, generally luminol (chemiluminescence) or ABTS (colorimetry) [2]. Researchers still are active in development of new peroxidase substrates, which would involve the use of other measurement techniques and would be more sensitive. Our interest is focused on a reaction of catalytic silver deposition. This reaction is well known and broadly used in system with horseradish peroxidase. We have investigated the possibility of adapting this reaction to systems containing peroxidase-mimicking DNAzymes and prospect of its practical use in combination with LSPR technique.
Despite the fact, that the reaction of silver deposition is well known, the mechanism and reaction conditions are still not fully understood. Our preliminary studies involved confirmation of this catalytic reaction in DNAzyme system by spectrophotometric technique. As a sensing platform we used an oligonucleotide composed of a TTTT(CH2)6SH linker and a PS2.M domain (3'GGGTAGGGCGGGTTGGG5'), which possesses proved high peroxidase activity in the complex form with hemin. On the basis of UV-Vis spectra, the wavelength of 406 nm was chosen for absorbance changes measurements that accompanied catalytic silver deposition. The results confirmed that DNAzyme with peroxidase activity catalyzed the reaction of silver deposition.
We decided to exploit this reaction in LSPR technique (Localized Surface Plasmon Resonance). The initial experiments confirmed binding between hemin and G-quadruplex forming sequence. We also observed the changes in light scattering of gold nanoparticles after silver deposition. The DNAzyme catalyzed silver deposition was much more effective comparing to the reference reaction without hemin. Those positive results give future prospects for developing bioassays by combination of DNAzyme catalyzed silver deposition and LSPR technique.
Acknowledgments:
This work was partially supported by Short Term Scientific Mission grant (COST Action MP0802)
[1] J. Kosman, B. Juskowiak, Anal.Chim.Acta 707 (2011) 7.
[2] P. Travascio, Y. Liu, D. Sen, Chem.Biol. 5 (1998) 505.
1Physik-Department, Technische Universität München, 85748 Garching, Germany; 2 Fakultät für Physik, Ludwig-Maximilians-Universität, 80539 München, German; 3Department of Physics and Astronomy, Ohio University, Athens, OH, 45701, U.S.A.
† Present address: Aalto University School of Science, FI-00076 Aalto, Finland
* anton.kuzyk@aalto.fi
For a long time molecular self-assembly has been considered as an attractive approach for the bottom-up construction of plasmonic structures from well-defined metal nanoparticles (NPs). The lack of experimental techniques for controlled and reproducible assembly of nanoparticles into predesigned complex structures has been one of the limiting factors in development of the field of NPs plasmonics. Although, DNA self-assembly has been widely used for arrangement of metal nanoparticles[1], up to date, most of the DNA guided assemblies of NPs showed rather limited optical functionality. An important recent development has been the introduction of DNA origami technique, through which the assembly of nanoscale objects of unprecedented complexity become feasible[2]. For DNA origami, hundreds of rationally designed "staple" oligonucleotides are hybridized to a long single-stranded DNA scaffold strand, which forces it to assume a specific two- or three-dimensional shape. The resulting objects are fully addressable by their DNA sequence, and this property can be utilized to decorate origami structures with nanocomponents in a unique, sequence-specific manner[3]. We used DNA origami for programmed assembly of complex plasmonic nanostructures with optical response that emerges as a collective effect from precise three dimensional arrangement of multiple nanoparticles in close proximity[4]. The design of our structures is illustrated in Figure 1A. Specifically, nine gold nanoparticles were arranged into right and left handed helices. The response of helical structures to an electromagnetic field is associated with the effect of circular dichroism (CD), which denotes the differential absorption of left- and right-handed circularly polarized light. For left handed helical arrangements of NPs, CD signal with a characteristic bisignate peak-dip shape is expected (Fig. 1C and 1D), where the peak is centred around the plasmon resonance[5]. Right-handed helices are expected to produce a vertically mirrored dip-peak signal. CD response can be dramatically increased by increasing the particle size through electroless deposition of metal from solution (Fig. 1D). In addition, our method can be easily modified to generate optical response at other wavelengths[4].
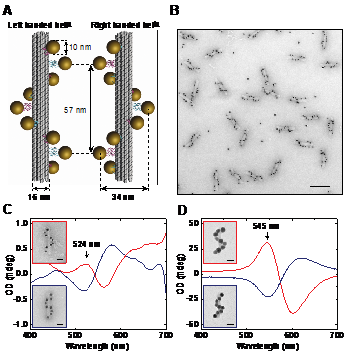
- Fig. 1: (A) In our design, DNA origami 24-helix bundles offer nine positions for attachment of NPs. (B) Transmission electron microscopy (TEM) image of assembled gold nanohelices (scale bar: 100 nm). (C) The CD spectrum of left-handed (red lines) and right-handed (blue lines) 10 nm gold particle helices. Color frames indicate the correspondence between the spectra and the nanohelices presented in the TEM images (scale bars: 20 nm). (D) The optical response increases dramatically and the CD peak shifts to longer wavelength with an increase of NPs size.
[1] Tan, S.J. et al. Nature Nanotechnol. 6, 268-276 (2011).
[2] Rothemund, P.W.K. Nature 440, 297-302 (2006); Douglas, S.M. et al. Nature 459, 414-418 (2009).
[3] Tørring, T. Chem. Soc. Rev. 40, 5636-5646 (2011).
[4] Kuzyk, A. et al. Nature, 483, 311-314 (2012).
[5] Fan, Z. & Govorov, A.O. Nano Lett. 10, 2580-2587 (2010).
National Museum of Natural History, CNRS UMR7196 / INSERM U565, 43 rue Cuvier, Paris, France lavelle@mnhn.fr
Through its local heterogeneities and transient structural changes resulting from chemical modifications and physical constraints imposed by numerous actors in vivo, chromatin dynamics influences (and is influenced by) DNA transcription, both at the initiation and elongation stages.
While transcription events often correlate with large chromatin movements, RNA polymerase access to its target sequence implies some nucleosome dynamics (potentially mediated by chromatin remodeling factors) and its subsequent tracking along the DNA template imposes some drastic topological and structural changes that propagate in the transcribed chromatin domain. Experiments and models aimed at deciphering the key features of chromatin dynamics and topology upon transcription will be presented.
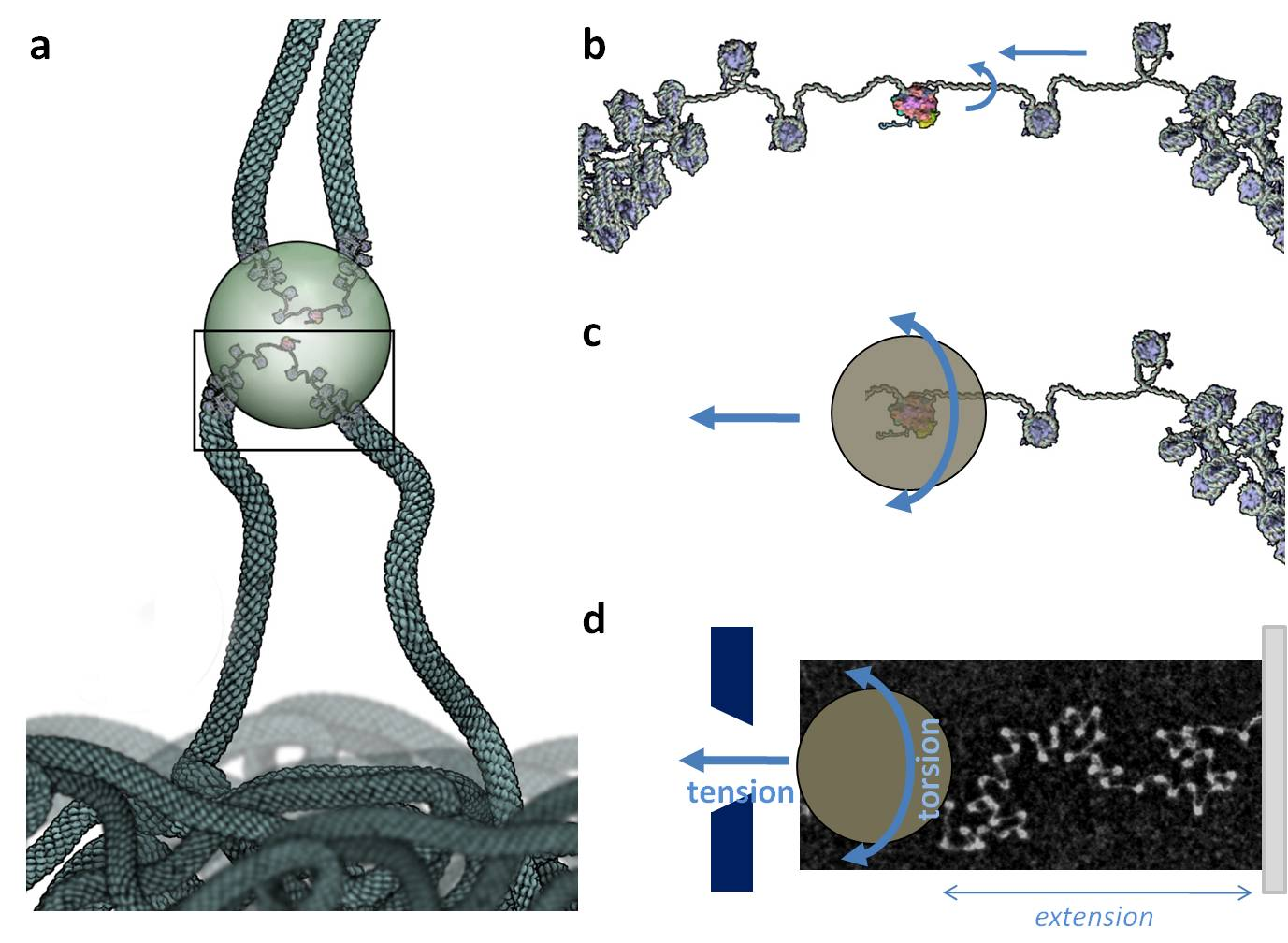
- Fig. 1: The transcription factory model and its mechanical consequences assessed in vitro. (a) Upon transcription, a chromatin loop expands out of a chromosome territory and polymerase pulls chromatin through the factory (potentially shared by other transcribed genes from adjacent chromosome territories). (b) Inside the factory, chromatin is pumped and screwed through the polymerase, undergoing both tension and torsion. (c,d) Both of these constraints can be reproduced in vitro by attaching a chromatin fiber between a glass slide and a paramagnetic bead which rotation and displacement is controlled by a pair of magnets. Reconstituted chromatin fibers from purified components (DNA and histones) are used for these studies, and usually checked by transmission electron microscopy (see picture in (d)) before use.
[1] Lavelle, C., Bancaud, A., Recouvreux, P., Barbi, M., Victor, J.M. And Viovy, J.L. .2011 Chromatin topological transitions. Prog Theor Phys 191:30-39.
[2] Recouvreux, P., Lavelle, C., Barbi, M., Conde e Silva, N., Le Cam, E., Victor, J.M. and Viovy J.L. 2011 Linker histone incorporation maintains chromatin fiber plasticity. Biophys J 100:2726-35.
[3] Lavelle, C., Recouvreux, P., Wong, H., Bancaud, A., Viovy, J.L., Prunell, A. and Victor, J.M. 2009 Right-handed nucleosome: myth or reality? Cell 139:1216-7.
[4] Lavelle, C. (2009). Force and torques in the nucleus: chromatin under mechanical constraints. Biochem Cell Biol 87:307-322.
[5] Lavelle. 2008 DNA torsional stress propagates through chromatin fiber and participates in transcriptional regulation. Nat Struct Mol Biol 15:123-5.
Institute of Photonic Technology, Albert-Einstein-Straße 9, 07745 Jena, Germany christian.leiterer@gmail.com
Bottom-up nanostructures such as semiconducting nanowires (NW) and noble metal nanoparticle (NP) can be synthesised reproducible at low cost. They can be used as highly sensitive optical or electrical sensors. Especially in the field of nano-electronics the integration of these low-cost nanostructures can be very difficult and are therefore cost intensive, preventing the development of a ready-for-market device.
Therefore a technique is required which allows the integration of bottom-up nanostructures into an electrical environment [1] which should work at nanoscale precision, apply sufficient contact to the electrode-nanostructure interface, but still can be performed at low cost. Here we present a technique, which fulfils these requirements.
The technique is based on a high frequency (1 kHz – 10 MHz) AC electrical field known as dielectrophoresis (DEP). We show that DEP-based integration of nanostructures (NW. NP) can be performed reproducibly with low contact resistance. Also, we show that the technique is parallelizable and can be monitored optically as well as electronically in order to control and characterize the integration process. Finally we will show that it is possible to establish optical- and (bio)-chemical sensors based on DEP-integrated nanostructures.
- Fig. 1: DEP integrated nanostructures (AuNp, SiNW) and their electrical response after the exposure to a chemical / optical signal. (AuNP, chemical sensing, left, SiNW, optical sensing, right)
[1] Wolff A, Leiterer C, Csaki A und Fritzsche W 2008 Front. Biosci 13 6834–6840
Chemical Nanoscience Laboratory, School of Chemistry, Bedson Building,
Newcastle University, Newcastle Upon Tyne, NE1 7RU, UK
e-mail: Shahrbanou.Moradpourhafshejani@newcastle.ac.uk, a.r.pike@newcastle.ac.uk*
DNA through programmable self-assembly, using the selective affinity of base pairs DNA strands has been used to form nanoscale materials. Numerous small molecules are known to bind into DNA through base pair intercalation. One way to use DNA to build functional nanomaterials is to accommodate functional groups with the aid of intercalators. In this poster, we present the synthesis of proflavine diazide derivatives as intercalators and building nano rings during click reaction and polymerisation at the presence of DNA.
1Institute of Photonic Technology, Albert-Einstein-Straße 9, 07745 Jena, Germany 2Institute of Physical Chemistry and Abbe Center of Photonics, Friedrich Schiller University, Jena, Helmholtzweg 4, 07745 Jena, Germany
susanne.pahlow@uni-jena.de, juergen.popp@uni-jena.de
DNA-chips, using fluorescence as standard detection method, are a very promising analytical tool in modern DNA diagnostics. However, there are certain drawbacks associated with this technique. For instance, difficulties during the hybridisation procedure on the chip surface might occur when longer DNA strands, for example from PCR products, are employed. Another problem arises from the use of fluorescence labelled targets, which can be subject to fluorescence quenching or photobleaching.
A fast, simple and cost efficient bioassay, combining magnetic beads and enzymatically generated silver nanoflowers, which allows easy optical detection for sequence specific DNA identification has been developed. The method has some distinct advantages over fluorescence based DNA detection on microarrays. As a matter of fact, hybridization can be carried out in solution and no fluorescence labels are required. Magnetic beads offer a high potential for numerous bioanalytical applications due to their unique properties, like their superparamagnetic cores and the nearly unlimited possibilities for surface modification with different functional groups. We employed magnetic beads for the immobilization of single-stranded DNA on their surface and performed a hybridization assay using enzyme labelled reporter DNA. After the hybridisation step, silver nanoflowers were generated on the magnetic beads by enzyme induced silver deposition. The specificity and sensitivity of our assay was evaluated using a PCR product of E. coli.
With enzymatically generated silver deposition on magnetic beads a DNA detection method without any additional hardware is given. It can be distinguished between positive and negative results by the naked eye. A major advantage of this experimental setup is the connection between sample preparation and detection in one step. The simple handling associated with magnetic beads has an enormous potential for point of care as well as on-site diagnostic where only a yes or no answer is necessary.
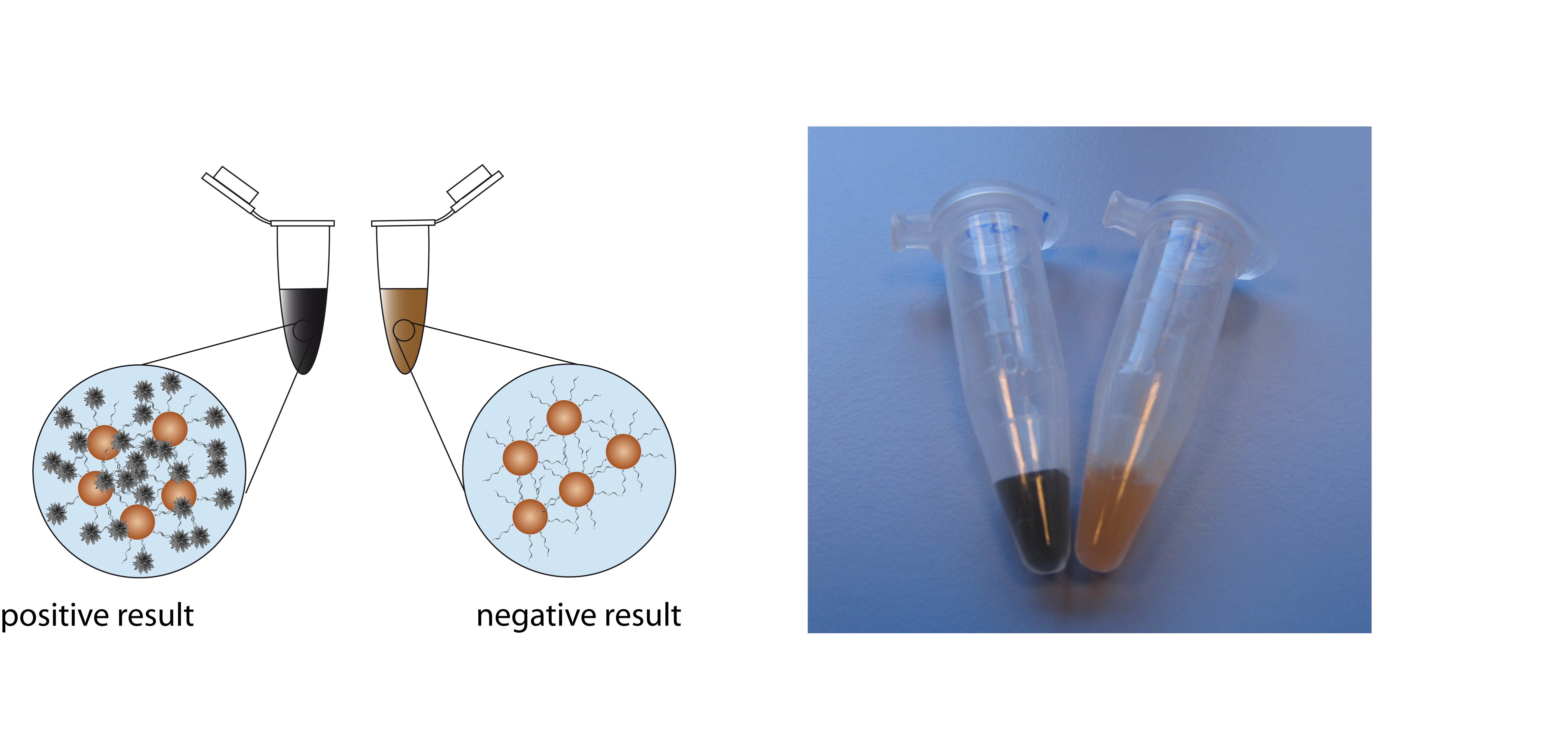
- Fig. 1: Positive and negative test results can be distinguished by the naked eye. The picture on the right side shows two samples, which either contained PCR product of E. coli (left tube) or a negative control (right tube).
Acknowledgement: Funding of the research project 'FastDiagnosis' from the Federal Ministery of Education and Research, Germany (BMBF) is gratefully acknowledged
aCBM S.c.r.l., SS14, KM 163,5 Basovizza, 34149 Trieste, Italy
bDip.to di Biologia e Protezione delle Piante, Università degli Studi di Udine, via delle Scienze 208, 33100 Udine, Italy
cIOM-CNR Laboratorio TASC, SS14, KM 163,5 Basovizza, 34149 Trieste, Italy
dInterdisciplinary Nanoscience Center (iNANO), Aarhus University, DK-8000 Aarhus, Denmark
eINBB Istituto Nazionale di Biostrutture e Biosistemi, Interuniversity consortium, Italy
§present address: Department of Physics, Middle East Technical University, Dumlupinar Bulvari 1, Universiteler Mahallesi, Cankaya, 06800 Ankara - Turkey
* Contact: Luca Piantanida. Tel.: +39 3402576493 E-mail adress: piantanida@iom.cnr.it
The DNA origami method allows the design and fabrication of arbitrary 2D and 3D shapes starting from one long single strand DNA with the simple introduction of short oligonucleotides which hold together the structure. Many examples of complex structures have been described in literature [1,2], while few results have been reported about actuation of movable origami [3]. We synthesized a DNA origami with a movable part that can be used for molecular recognition. It basically consists of one 100 nm diameter external ring that is linked at two opposite points to an internal movable disk. A single strand DNA filament is linked through two protruding staple strands placed at opposite points on the two halves of the internal disk. When a complementary ssDNA hybridizes with it, the new construct pulls the opposite sides of the disk which, as a consequence, folds on itself. The Atomic Force Microscopy technique is largely used for the visualization of DNA structures, and in particular of DNA origami structures. In this work we report the morphological and functional characterization of this particular DNA origami structure. The switch mechanism was thoroughly investigated and we demonstrated a proper design to obtain a reproducible switch reversibility. Furthermore we evaluated and demonstrated that the origami switch maintains its functionality also when the structure is placed on a solid substrate, allowing the integration of our switch within solid state based devices [4]. In order to appreciate the directionality of the switch movements we planned to analyze our device with other techniques. The FRET systems and the plasmon resonance technique could help to sense the distance between the different movable parts of the origami in a more sensitive and controllable way. We want to modify our DNA structure inserting, in a defined position, gold nano-particles making it suitable for FRET and plasmon resonance technique. In the early future we will create a new DNA structure to calibrate the potential of these techniques.
[1] Rothemund, P. W. (2006). "Folding DNA to create nanoscale shapes and patterns." Nature 440(7082): 297-302.
[2] Andersen, E. S., M. Dong, et al. (2008). "DNA origami design of dolphin-shaped structures with flexible tails." ACS Nano 2(6): 1213-1218.
[3] Andersen, E. S., M. Dong, et al. (2009). "Self-assembly of a nanoscale DNA box with a controllable lid." Nature 459(7243): 73-76.
[4] Marini, M., Piantanida, L. et al. (2011). "A revertable, autonomous, self-assembled DNA-origami nanoactuator." Nano Lett., 2011, 11 (12), pp 5449–5454.
Laboratory of Applied and Environmental Microbiology, IRTA – Torre Marimon, Torre Marimon, Caldes de Montbui, Spain. francesc.prenafeta@irta.cat
DNA technology has revolutionized fundamental and applied sciences in the latter decades. Environmental biotechnology has also been benefiting from these developments, and the characterization of the microbial community structure and dynamics is progressively changing the traditional "black box" bioreactor view. The understanding of the underlying interactions between the microbiota and the environment (directly related to operational parameters in engineered systems) represents an important source of information for an optimized bioreactor operation. Monitoring the microbial composition might hint the implementation of strategies that promote desirable and stable consortia of microorganisms (e.g. positive syntrophic interactions, specialized biodegraders) while preventing the presence of detrimental species (e.g. pathogens, unwanted trophic competitors, biodeterioration agents).
We are providing a set of customized protocols aimed to the characterization of whole microbial communities in different kinds of bioprocesses applied to environmental technology, such as anaerobic digesters for the treatment of organic wastes [3], sequential nitrification-denitrification batch reactors [2], biofilters for the decontamination of air polluted with volatile organic compounds [4], and even in the bioremediation soils and aquifers containing emerging pollutants [1]. This molecular toolbox is based, as the first step, on the amplification by PCR of a particular DNA fragment from a mixture of organisms present in environmental samples. Ribosomal genes are usually targeted because of their high phylogenetic resolution while allowing the use of domain-specific primers (Eubacteria, Archaeobacteria, and microbial Eukarya such as fungi). Also, a wide range of functional genes linked to specific trophic modes (e.g. methanogenesis, dehalogenation, biological nitrogen cycle) might be amplified selectively by PCR. Mixed amplicons can then be resolved by Denaturing Gradient Gel Electrophoresis (DGGE), which generates a genetic fingerprint of the microbial community. Individual DNA sequences are depicted as specific bands that can be excised and sequenced to identify the dominant members of the microbial population. These profiles can be digitalized and parameterized in a numerical form, allowing subsequent multivariate analysis and statistical correlation with environmental parameters. The easy comparison of the community structure of several samples facilitates the assessment of environmental gradients in time and/or space. However, only the dominant populations are represented and, in highly biodiverse samples, the obtained DGGE profiles might be too fuzzy for interpretation.
On the other hand, quantitative PCR (qPCR) represents an ideal complement to the more qualitative PCR-DGGE. The quantification of specific genes providing both functional and phylogenetic information allows precise estimations on the evolution of specific microbial ratios (fungal/bacterial, arquea/bacteria, methanogens/arquea, etc.) which are relevant to the system behaviour [4]. Finally, the integration of both, qualitative and quantitative molecular data can be accomplished by means of high throughput sequencing tools such as pyrosequencing. This emerging technique has prompted the in-depth characterization of the microbial biodiversity and even the whole description of metagenomes. The resulting sequence datasets are very large and need to be handled by specific bioinformatic pipe-lines for automated phylogenetic assignment and biodiversity estimation. We are currently applying the pyrosequencing of tagged 16S rRNA gene amplicons in evaluating the impact of different waste pre-treatment strategies on the microbial community structure of anaerobic digesters treating lipid rich wastes.
In summary, the implementation of a combined approach based on different PCR-based techniques (DGGE, qPCR, and pyrosequencing) provides information that is useful for the "prove of concept" in new bioreactor design, but might also assists operators of industrial plants on the early diagnoses of potential system malfunction. Last, but not the least, the molecular data generated in the lab could also be used in the set up and validation of the performance of advanced miniaturized biosensors prior to, or during, their application in-situ.
[1] De Weert, J., Viñas, M., Grotenhuis, T., Rijnaarts, H., & Langenhoff, A. (2011). Degradation of 4-n-nonylphenol under nitrate reducing conditions. Biodegradation 22:175-187.
[2] Magrí, A., Guivernau, M., Baquerizo, G., Viñas, M., Prenafeta-Boldú, F.X., & Flotats X. (2009). Batch treatment of liquid fraction of pig slurry by intermittent aeration: process simulation and microbial community analysis. Journal of Chemical Technology & Biotechnology 84:1202-1210.
[3] Palatsi, J., Illa, J., Prenafeta-Boldú, F.X., Laureni, M., Fernandez, B., Angelidaki, I., & Flotats, X. (2010). Long-chain fatty acids inhibition and adaptation process in anaerobic thermophilic digestion: batch tests, microbial community structure and mathematical modelling. Bioresource Technology 101:2243-51.
[4] Prenafeta-Boldú, F.X., Guivernau, M., Gallastegui, G., Viñas, M., de Hoog, G.S., Elías, A. 2012 Fungal/bacterial interactions during the biodegradation of TEX hydrocarbons (toluene, ethylbenzene and p-xylene) in gas biofilters operated under xerophilic conditions. FEMS Microbiology Ecology (In press).
Technische Universität München, Chemistry Department & Walter Schottky Institute, Garching, Germany
rant@tum.de
The 'switchSENSE' principle is based on an electrically actuated bio-interface. Short end-tethered DNA molecules are driven to oscillate (switch their conformation) on microelectrodes by the application of AC potentials. The induced molecular motion is monitored in real-time by fluorescence energy transfer. It not only reveals the mere presence of biomolecular targets on the sensor surface, but also permits the analysis of the target molecule size, shape, and charge in a single experiment.
I will describe how target molecules are swayed through the solution atop of DNA levers and discuss how kon, koff, KD values are determined from the DNA's switching behaviour. The molecular size of the target is inferred from molecular dynamics measurements, which are analysed with a theoretical model that takes entropy, charging effects, and hydrodynamic friction into account. The implications for a novel type of protein biochip will be discussed.
University of Rome, Tor Vergata, Via della Ricerca Scientifica, 00133, Rome, Italy, francesco.ricci@uniroma2.it
The development of rapid, low cost, point-of-care approaches for the quantitative detection of transcription factors (TFs) and other clinically relevant biomarkers such as antibodies would drastically impact healthcare, narrowing the gap between diagnosis and treatment, and improving the penetration of modern diagnostics into the developing world. Unfortunately, current methods for the quantitative detection of proteins and antibodies including ELISAs and western blots, are typically complex, multiple step processes requiring well trained technicians in well equipped laboratories. Here, in contrast, I report the design of versatile, DNA-based electrochemical and optical switches for the quantitative, single-step detection of specific TFs that we termed "Transcription Factor Beacons". The TF beacon strategy is inspired by naturally occurring biomolecular switches that employ binding-induced structural change to signal the presence of a specific molecular target or its functioning. Using these sensors we demonstrated the rapid, quantitative detection of physiologically relevant, low-nanomolar concentrations of transcription factors directly in complex clinical samples. The TF beacons we describe in this presentation represent a versatile new class of binding-activated probes for the monitoring of specific DNA binding activity. These new probes achieve biologically relevant specificities and detection limits. Critically, because their signalling is induced only by the formation of a highly specific probe–target complex, such conformation-linked sensors work well even when deployed in complex environments. Given these attributes, the TF beacons may prove of significant utility in a range of applications, including drug screening, cancer diagnostics, and developmental biology, where interest in the quantitative regulation of TFs is rapidly growing. Using a similar approach we also demonstrate the proof-of-principle for electrochemical DNA-based switches that respond to specific antibodies. We find that theses switches can allow the detection of specific antibodies quantitatively, selectively, rapidly (< 5 minutes), and achieve low nanomolar detection limits even when challenged directly in complex clinical samples.
1 Institute of Photonic Technology, Albert-Einstein-Straße 9, 07745 Jena, Germany
2 Institute of Physical Chemistry and Abbe Center of Photonics, Friedrich Schiller University Jena, Helmholtzweg 4, 07743 Jena, Germany
Barbara.Seise@ipht-jena.de, Juergen.Popp@ipht-jena.de
Metal nanoparticles feature plasmonic activities in the UV to NIR range due to their different shapes, sizes and materials resulting in a wide range of application fields. They are implemented in biochip technologies, biosensors, molecular plasmonics and in biophotonics. Micro structured optical fibers serve as innovative optical devices when prepared with well-defined plasmonic layers of metal nanoparticles using different readout techniques.
A suspended core fiber with three holes containing metal nanoparticles as plasmonic structures on the internal capillary wall was produced. Implementation of self-assembling monolayer techniques enables plasmonic structures with a homogenous distribution of metal nanoparticles inside the fiber. These micro structured optical fibers serve as sensors for biomolecule interactions inside the fiber applying surface enhanced Raman spectroscopy (SERS) or localized surface plasmon resonance (LSPR) sensing for readout.
Within this contribution, the application of the SERS technique for analytical devices combined with the micro structured optical fiber is introduced. Investigating the nanoparticle modified fiber as SERS sensor, crystal violet was applied in a first experiment. Raman spectra of crystal violet filled fibers with and without nanoparticles were recorded. The fiber without nanoparticle shows a weak crystal violet spectrum while the fiber filled with nanoparticle creates a strong crystal violet spectrum due to the SERS effect. Further, DNA hybridization was performed inside the fiber using Cy3-labeled target DNA. Within the fiber, Cy3-label could be recorded via SERS.
In conclusion, a suspended core fiber with three holes containing metal nanoparticles as plasmonic structures on the internal capillary wall is presented as innovative optical device allowing both LSPR and SERS readout. Implementation of self-assembling monolayer techniques enables plasmonic structures with a homogenous distribution of metal nanoparticles. These micro structured optical fibers can be used as sensors for DNA sensing as demonstrated.
We thank BMBF project NAWION (FKZ: 16SV5386K, V4MNI014) for financial support.
A. Csáki, S. Berg, N. Jahr, C. Leiterer, T. Schneider, A. Steinbrück, D. Zopf, W. Fritzsche, in: P. E. Chow (Ed.): "Gold Nanoparticles: Properties, Characterization and Fabrication", p 245-261 , Nova Science Publishers, 2010.
K. Hering, D. Cialla, K. Ackermann, T. Dörfer, R. Möller, H. Schneidewind, R. Mattheis, W. Fritzsche, P. Rösch, J. Popp, Analytical and Bioanalytical Chemistry, 390, 1, 113-124 (2008).
A. Csáki, F. Garwe, A. Steinbrück, G. Maubach, G. Festag, A. Weise, I. Riemann, K. Konig, W. Fritzsche, Nano Lett, 7, 247-253 (2007).Wirth, J., Garwe, F., Hähnel, G., Csaki, A., Jahr, N., Stranik, O., Paa, W. & Fritzsche, W. 2011 Plasmonic nanofabrication by long-range excitation transfer via DNA nanowire. Nano Letters 11
Environmental Toxicology Group, CSIR-Indian Institute of Toxicology Research, Lucknow-226 001, INDIA; * CSIR-National Physical Laboratory, New Delhi-110012, INDIA,
rishi@iitr.res.in
The surface and potable water quality has declined due to rapid industrialization and population growth. The microbiological quality of surface waters of riverine systems is adversely affected by several point and non-point sources of pollution. Further, untreated surface waters are inadvertently consumed for drinking and various household tasks in India making the public vulnerable to water-borne diseases and outbreaks. Our studies have shown the distribution of species diversity, dissemination of antimicrobial resistance and virulence-markers in water quality 'indicator' organisms such as Escherichia coli and Enterococci with respect to rural-urban landscape along river Ganga and its tributaries as well as potable water. We have developed q-PCR and Nano-Probes for detection of pathogens like Enterotoxigenic Escherichia coli, Enterohemorrhagic Escherichia coli and Vancomycin Resistant Enterococci and Salmonella in low doses in water and food. The molecular probe based studies have demonstrated the prevalence of ETEC and VRE in aquatic flora and cultivated leafy vegetables. This flora serves as environmental reservoir for the multi-antimicrobial agent resistant pathogens along landscapes of gangetic riverine system. In the New-INDIGO project AQUATEST, the prior art in our studies intends to foster development and validation of novel surveillance tools and remediation strategies to protect public health.
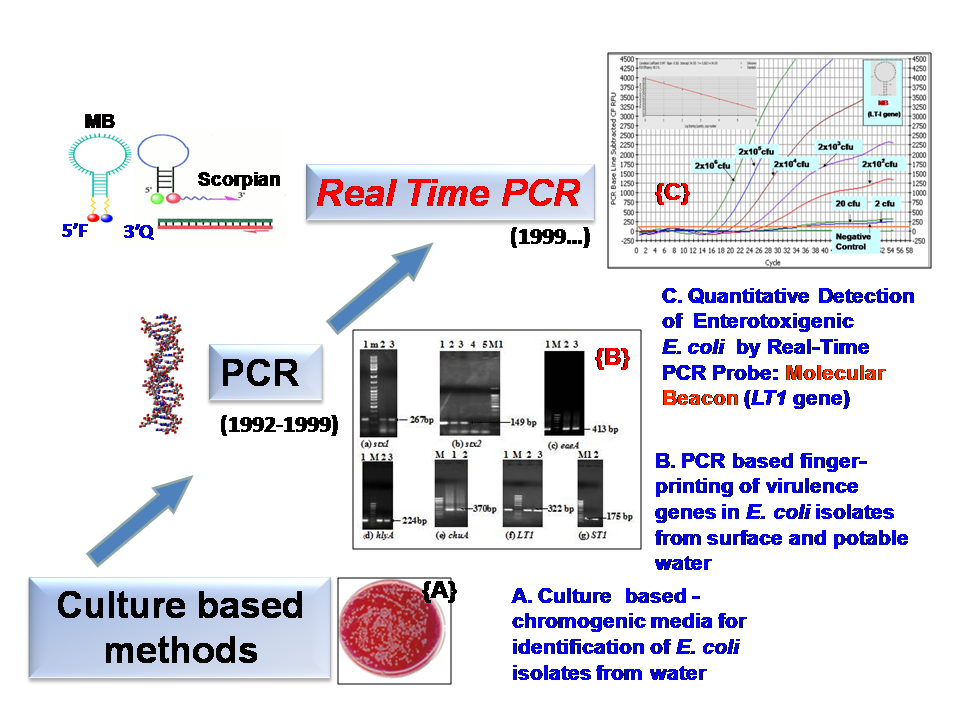
- Fig. 1: Detection of Water-Borne Pathogens
[1] Singh G., Vajpayee P., Ram S., and Shanker R. 2010. Environmental Reservoirs for Enterotoxigenic Escherichia coli in South Asian Gangetic Riverine System. Environmental Science & Technology 44: 6475–6480.
[2] Jyoti A., Pandey P., Singh S.P., Jain S.K. and Shanker, R. 2010. Colorimetric Detection of Nucleic Acid Signature of Shiga Toxin Producing Escherichia coli using Gold Nanoparticles. Journal of Nanoscience and Nanotechnology 10:1-5
[3] Jyoti, A., Ram, S., Vajpayee, P., Singh, G., Dwivedi, P.D., Jain, S.K., and Shanker, R. 2010. Contamination of surface and potable water in South Asia by Salmonellae: Culture-Independent quantification with Molecular Beacon real-time PCR. Science of the Total Environment 408: 1256–1263.
[4] Lata P., Ram S., Agrawal M. and Shanker R. 2009. Real-Time PCR for Rapid, Detection of vanA Gene in Surface Waters and Aquatic Macrophyte by Molecular Beacon Probe. Environmental Science & Technology 43: 3343-3348.
Nanoscience Center, 1Department of Physics and 2Department of Biological and Environmental Science, University of Jyväskylä, P.O. Box 35, FI-40014 Jyväskylä, Finland
3Institute of Biomedical Technology, University of Tampere and Tampere University Hospital, FI-33014 Tampere,Finland boxuan.shen@jyu.fi
Despite the rapid development of DNA nanostructures which can serve both as template and functional components for nanoscale devices, the precise spatial control and connection of these microscopic DNA composed nanostructures to macroscopic world still remains a huge challenge.
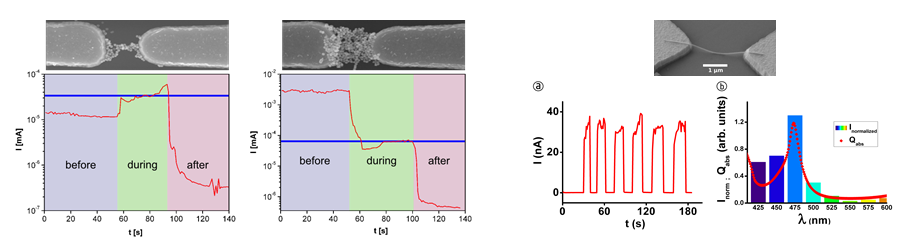
We present a method for controlled connection of gold electrodes with dsDNA molecules (locally on a chip) by utilizing DNA polymerase to elongate single-stranded DNA primers attached to the electrodes [1]. Thiol-modified oligonucleotides are directed and immobilized to nanoscale electrodes by means of AC dielectrophoretic (AC-DEP) trapping, and extended in a thermo-cycle, finally forming a complete dsDNA molecule bridging the gap between the electrodes (Fig. 1(b)). The presented technique can serve as a bridge between novel DNA nanotechnology and common molecular biology methods, while resolving also the problem of the precise spatial control. Furthermore, by utilizing multi-electrode geometries, it is possible to immobilize primers only to desired electrodes (Fig. 1(a)), thus enabling a specific and sequence depended growing of dsDNA between the chosen electrodes and thus sophisticated connection systems for detection and sensor applications as well as for molecular electronics.
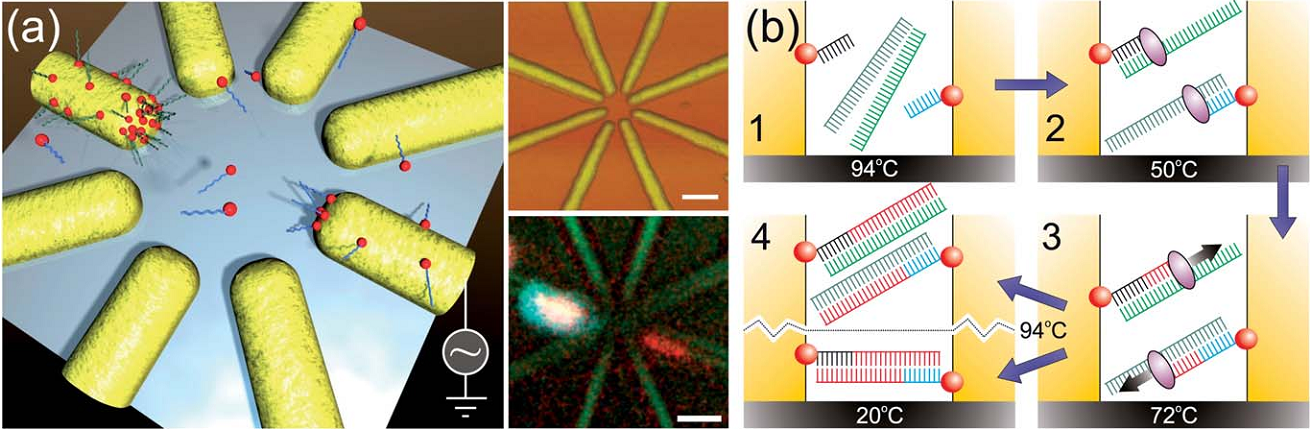
- Fig. 1: (a) - Left: a schematic view of the electrode-specific dielectrophoretic trapping of different thiol-modified primers with a multielectrode system. Upper right: an AFM image of the multielectrode structure. The scale bar is 100 nm. Lower right: a confocal microscope image of 40 nt long primers labelled with different dye molecules [Cy3 (red) and Cy5 (blue)] separately trapped and immobilized to the opposite electrodes. The scale bar is 1 mm. (b) - A series of schematic cross-sectional images presenting the thermo-cycle of the polymerase growth method (the final outcome can be either a dsDNA molecule bridging the two electrodes or pair with a template strand forming a one-end-immobilized dsDNA) [1].
[1] Linko, V., Leppiniemi, J., Shen, B.X., Niskanen, E., Hytönen, V.P., & Toppari, J.J. 2011. Growth of immobilized DNA by polymerase: bridging nanoelectrodes with individual dsDNA molecules. Nanoscale 3(9), 3788-3792
Institute of Photonic Technology, Albert-Einstein-Straße 9, 07745 Jena, Germany
christine.steinbach@ipht-jena.de, juergen.popp@ipht-jena.de
An improvement towards personalized cancer therapy is the selection of therapeutics based on sensitivity biomarkers. Circulating tumour cells (CTCs) are shed away from a primary tumour, stream via the peripheral blood to distant sites and can form metastases. Besides the CTCs' potential of metastasis, these cells can be used as a tool for clinical management of cancer patients and treatment monitoring. In this context, the evaluation of the mutation status of several cancer related genes in CTCs is an important issue. For instance, colon cancer patients with mutated KRAS (Kirsten rat sarcoma) do not respond to EGFR-inhibitors (epidermal growth factor receptor) [1]. Accordingly, only one nucleotide exchange has crucial impact on therapy efficacy.
Searching for such single nucleotide polymorphisms (SNPs) is an important challenge. To that goal, an in-house developed chip based system for a rapid and multiplexed readout is tested. This approach is based on the hybridization of immobilized capture DNA with biotin labelled sample DNA. The biotin binds to streptavidin conjugated horseradish peroxidase in a gap between micro structured electrodes. Subsequently, an enzyme induced silver deposition occurs which bridge the gap by a conductive layer [2, 3].
For future applications, the biochip should be integrated in a microfluidic point-of-care system. This could provide a fast and cost-effective diagnosis of specific cancer related mutations in CTCs.
Acknowledgments
Funding of research project "MiNa-CTC" by the Federal Ministry of Education and Research (BMBF), Germany is gratefully acknowledged.
[1] Martini M., Vecchione L., Siena S., Tejpar S., Bardelli A., 2011: Targeted therapies: how personal should we go?, Natural Review Clinical Oncology 9(2), 87-97.
[2] Seise, B., Brinker A., Kretschmer R., Schwarz M., Rudolph B., Kaulfuß T., Urban M., Henkel T., Popp J, Möller R., 2010: Chip-based detection system for the on-site analysis of animal diseases, Engineering in Life Sciences, 11(2), 148-156
[3] Schüler, T., Kretschmer R., Jessing S., Urban M., Fritzsche W., Möller, R., Popp J., 2009: A disposable and cost efficient microfluidic device for the rapid chip-based electrical detection of DNA, Biosensors Bioelectronics, 25, 15-21
1Nanoscience Center, Department of Physics, P.O.Box 35, FI-40014 University of Jyväskylä, Finland
2Institute of Biomedical Technology, University of Tampere and Tampere University Hospital, FI-33014 Tampere, Finland
3Institute of Photonic Technology, Albert-Einstein-Straße 9, 07745 Jena, Germany kosti.t.o.tapio@jyu.fi
Due to its superior self-assembly properties DNA is a very promising molecule for ever downscaling nano- and molecular electronics. During recent decades number of different self-assembled DNA-structures has been developed, e.g. triple crossover (TX) tiles and DNA origamis. These structures can serve as a scaffold for nanoscale electronic components, e.g. single electron transistor (SET).
Here we utilize a self-assembled DNA-structure consisting of two different TX-tiles (A and B) forming a definite chain of three tiles, i.e. BAB. These self-designed structures serve as a scaffold for gold nanoparticles (AuNPs) functionalised with thiolated DNA oligonucleotides: Each tile has a sticky end for attaching an AuNP so that the fully assembled structure consists of a sequence of three AuNPs which form the needed metallic islands for SET. The BAB structures were formed by heating the solution containing all the DNA oligonucleotides forming the BAB to 90°C and annealing it down to room temperature. The BAB-AuNP conjugates were fabricated similarly by slowly annealing the mixture from 45°C down to 21°C. Subsequently, the conjugation was verified by AFM imaging. Furthermore, BAB-AuNP conjugates were trapped between nanoelectrodes for electrical measurements by using dielectrophoresis. AFM imaging was utilized in obtaining the optimal parameters for trapping. The IV-curves of trapped BAB-AuNP conjugates were measured in room temperature and at 4.2K. However, the conductivity was extremely low at any measured temperature, most probably due to a too large gap between AuNPs, not allowing any significant electron tunneling. Thus, the AuNPs of the conjugated and trapped structure were further grown by using a commercial electrochemical gold enhancer kit (Nanoprobes – cat. # 2113). Growth of 5-8 nm was obtained via dilution of the solutions, and verified again by AFM imaging. After that the IV measurements revealed clear Coulomb Blockade on couple of samples. The dependency of the blockade on the temperature was verified at several temperatures between 4.2 K and 100 K. Future measurements will concentrate on IV-curve dependence
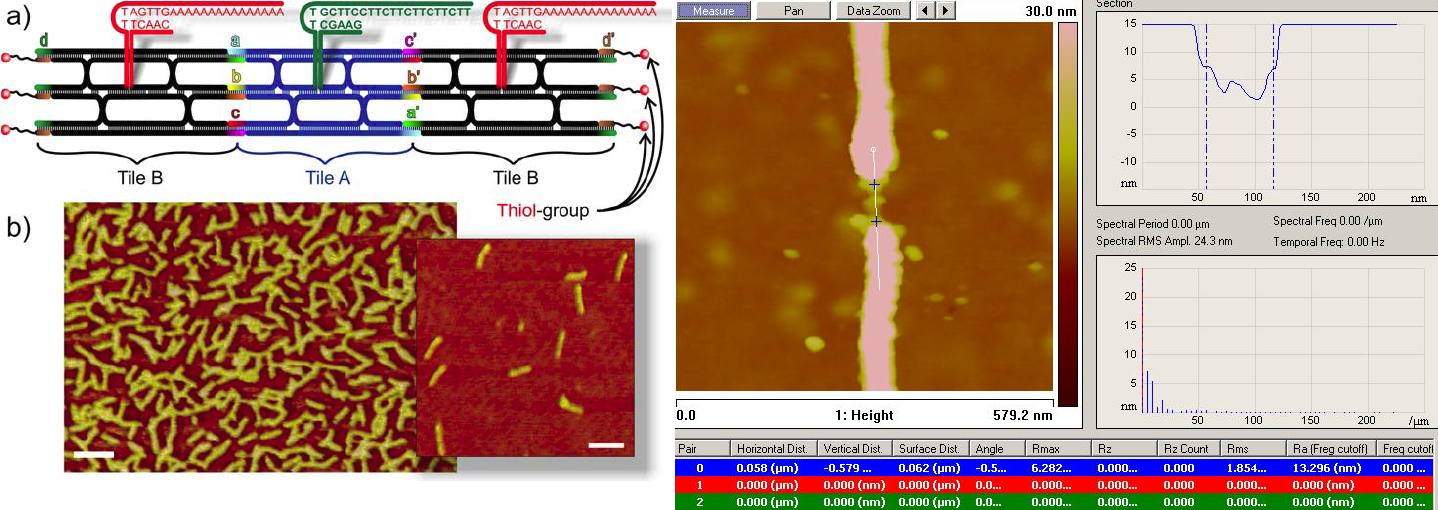
- Fig. 1: a) - Schematic view of the self-assembled DNA structure (BAB) with sticky ends for functionalized AuNPs . b) - AFM image of the formed BAB structures without AuNPs. The scale bar is 100nm. c) - AFM image of a trapped conjugate structure: Bumps in the height profile are the AuNPs of the structure.
[1] Linko, V., Leppiniemi, J., Paasonen, S.-T., Hytönen, V.P., & Toppari, J.J. 2011 Defined-sized DNA triple crossover construct for molecular electronics: modification, positioning and conductance properties. Nanotechnology 22, 275610
1Nanoscience Center, Department of Physics, P.O.Box 35, FI-40014 University of Jyväskylä, Finland
2Institute of Biomedical Technology, University of Tampere and Tampere University Hospital, FI-33014 Tampere, Finland
3Department of Applied Physics, P.O.Box 5100, FI-02015 Helsinki University of Technology, Finland
j.jussi.toppari@jyu.fi
The most crucial part in the realization of functional and multicomponent molecular scale nanodevices is to find a suitable scaffold for the patterning. For the moment, DNA has been proven to be a very flexible and promising molecule for this, mostly due to its superior self-assembly properties. In this respect, the electrical properties of DNA-scaffolds are also of a great interest. In addition, to be able to measure the electrical properties or otherwise realize the full potential of the bottom-up DNA-nanotechnology, one needs to position these constructs on the chip in a controllable way. We have shown that dielectrophoresis (DEP) can be efficiently utilized to trap DNA molecules and self-assembled structures, such as DNA origami [1].
Here we have utilised DEP to trap rectangular DNA origamis and three-tile-long defined TX-tile constructs between the nanoelectrodes, and characterized the electrical conductivity of them. Both DC and AC characteristics of individual trapped structures, surrounded by atmosphere with different humidity levels, have been fully analyzed by impedance spectroscopy and detailed equivalent circuit modelling, describing the conductivity mechanisms of the structure [2,3]. The results revealed that the nature of the conductivity is a combination of an ionic diffusion and Ohmic conductivity. It also scales with the volume of the structure pointing towards water/humidity induced conductivity along the DNA helices, most possibly by the layer of polarised water molecules sheathing the DNA. Further measurements with even more controllable structures are on the way.
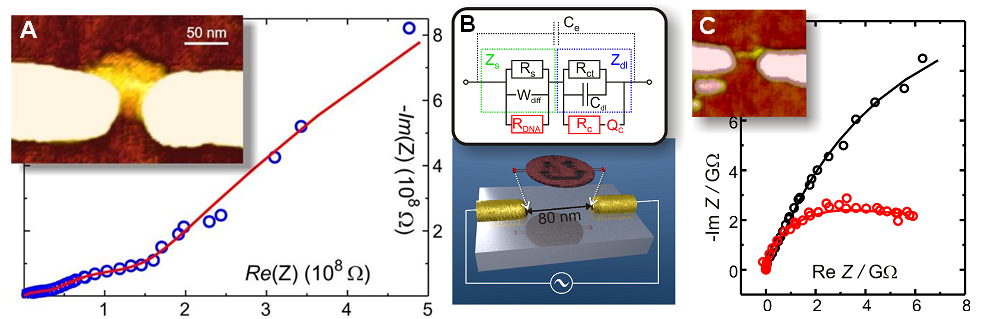
- Fig. 1: A - Inset: AFM Image of a rectangular DNA origami trapped and immobilized between nanoelectrodes. Graph: AC impedance spectroscopy data measured from the structure (circles) and the response of a fitted equivalent circuit (line). B – Upper: Used equivalent circuit model. Lower: Schematic view of trapping of a DNA origami. C - Inset: AFM Image of a TX-tile structure trapped and immobilized between nanoelectrodes. Graph: AC impedance spectroscopy data measured from these structures (circles) and the response of a fitted equivalent circuit (lines).
[1] Kuzyk, A., Yurke, B., Toppari, J.J., Linko, V., & Törmä, P. 2008 Dielectrophoretic trapping of DNA origami. Small 4(4), 447
[2] Linko, V., Paasonen, S.-T., Kuzyk, A., Törmä, P., & Toppari, J.J. 2009 Characterisation of the Conduc¬tance Mechanisms of the DNA Origami by AC Impedance Spectroscopy. Small 5(21), 2382
[3] Linko, V., Leppiniemi, J., Paasonen, S.-T., Hytönen, V.P. & Toppari, J.J. 2011 Defined-sized DNA triple crossover construct for molecular electronics: modification, positioning and conductance properties. Nanotechnology 22, 275610
Nanoscience Center, Department of Physics, P.O. Box 35, FI-40014 University of Jyväskylä, Finland
Detection of any DNA sequences of interest, e.g., for harvesting of microbials, pathogens or genes, is always based on the superior capability of single stranded DNA to recognize its complementary strand and thus form a double helix. For successful detection one needs some means to organize DNA catcher strands complementary to the target strand, and a way to generate readable signal after the target has hybridized with the catcher. Traditionally the signal generation has been based on optical readout, e.g., via (localized) surface plasmon resonance, FRET or regular fluorescence. However, these methods need rather complicated devices for reading or often require substantial amount of target to generate a readable signal. Quicker and easier to implement would be to use electrical readout, which at its best would generate a signal even from single hybridized target. For this purpose the electrical properties of DNA is of a great interest.
Since the direct conductance measurements are feasible only for very short DNA helices, some more advanced methods are needed for electrical detection of longer target sequences. Due to this more information is needed from the nature of the DNA conductivity. We have trapped single DNA molecules or more complicated self-assembled constructs between nanoelectrodes, and characterized the nature of their electrical conductivity by both DC and AC impedance spectroscopy. Also, methods to selectively enhance the conductivity of hybridized targets later on the second step, e.g. by metallization or ionic modification, have been investigated. Also, fabrication of electrically active DNA marker attaching and activating only when the desired target is present, could be designed. On the other hand, DNA self-assembly could be utilised to build an electrical detector for other purposes. For example, we have utilized self-assembly of so-called TX-tiles to fabricate a single electron transistor. These devices have been shown to be sensitive even to a fraction of electron charge.
Institute of Photonic Technology, Albert-Einstein-Straße 9, 07745 Jena, Germany stefanie.treppner@ipht-jena.de
The quality of drinking water has to meet certain requirements like purity, suitability for consumption and absence of contaminations with harmful substances and pathogens [1]. The current standard for detection of pathogen water-microorganisms is a cultivation process over several days [2], which inhibits an immediate treatment of the contamination. For this reason the project "Risk Management In Drinking House Installation (RiMaTH)" combines already established methods based on molecular biological techniques for a faster detection.
This goal includes the development of an integrated system for a fast detection, classification and activity analysis of bacterial contaminations as well as the advice on regeneration purposes.
The Institute of photonic Technology offers different nucleic-acid-based chip-technologies, which will be further developed for an on-site analysis. For this purpose the molecular biological method polymerase chain reaction (PCR) is used. BioSolutions Halle GmbH designs the specific oligonucleotides which are needed for the PCR and subsequent detections.
After performing the PCR on a PCR-chip, developed by the IPHT [3], two approaches for the detection of pathogens want to be realized: On one hand the PCR-Chip wants to be combined with the electrical DNA-Chip, which gives the opportunity to detect the PCR-products by specific probes (fig. 1 C). On the other hand the produced PCR-products will be detected on-line with fluorescence markers. This enables a real-time detection of PCR, high resolution melting curve analysis, homogenous hybridization assays and quantification (fig. 1 D). Furthermore the usually used gel electrophoresis for detection of PCR-products is not needed. Both approaches will be prepared on DNA and RNA base and normal PCR as well as multiplex-PCR should be conducted.
The comparison of the previous described detection methods with reference methods (cultivation, conventional quantitative PCR), supply of samples and advice of building operators are the tasks of the Federal Environment Agency (UBA) (fig. 1 A and B).
As industrial partner Analytik Jena AG has a broad level of provision of infrastructure, which gives the opportunity to realize a laboratory prototype of the chip-based technologies.
A proof-of-concept study which uses a culture-free method with Raman spectroscopy for a detection of pathogens is contributed by the Institute of Physical Chemistry (IPC) at the University of Jena (fig. 1 E). This also includes the establishment of a Raman-database for an automatic identification of pathogens. In contrast to the chip technologies no mobile but uncomplicated and very fast measurements are possible.
The project is supported through the Federal Ministry of Education and Research (BMBF, FKZ 02WRS1276A).
[1] Umweltbundesamt (2011): Bericht des Bundesministeriums für Gesundheit und des Umweltbundesamtes an die Verbraucherinnen und Verbraucher über die Qualität von Wasser für den menschlichen Gebrauch (Trinkwasser) in Deutschland.
[2] Bundesamt für Justiz (2011): Verordnung über die Qualität von Wasser für den menschlichen Gebrauch (Trinkwasserverordnung – TrinkwV).
[3] Felbel et al. (2007): DNA-Vervielfältigung mit Mirkochip-Thermocyclern. BIOforum 11, 36-38.
Chemistry Department, Technische Universität München, Lichtenbergstrasse 4, 85748 Garching, Germany
Walter Schottky Institute, Technische Universität München, Am Coulombwall 3, 85748 Garching, Germany;
valentina.villa@tum.de
Electrically actuated DNA layers can be used for label-free detection of protein-protein interactions in real-time on a chip. This technology combines electrical modulation of surface-tethered ds-DNA probes with an optical method to observe the dynamics of DNA conformational changes.
Here we present a comparative antibody affinity study. We determine the kinetic rate constants and affinities of four different monoclonal antibodies against the human enzyme Carbonic Anhydrase 1 (hCA1), which is coupled to a switchable DNA layer (Fig. 1-A). At first, we describe a suitable functionalization protocol by hybridizing preformed single stranded DNA layers with purified hCA1/complement-strand-DNA mono conjugates. Those stable conjugates are prepared by reacting benzaldehyde-modified DNA single strands with hydrazinopyridine-modified hCA1, which yields a bisaryl hydrazone covalent bond complex. Particular emphasis is put on the preparation of mono-conjugates, i.e., a 1:1 stoichiometry of DNA and protein. The presence of hCA1 on the surface is identified by a molecular dynamics measurement (Fig. 1-B). By loading nanomolar concentrations of each antibody on hCA1 modified- and reference-layers, the binding and unbinding kinetics are monitored optically in real time. Again, the dynamic switching response of the electrically modulated layers is used to identify the binding state of molecules, since the increase in hydrodynamic drag slows the switching movement (Fig. 1-B/C).
This chip-platform is suitable for multiplexed analysis, since the micro-electrodes can be individually addressed with different protein/DNA conjugates. This enables high throughput assays for antibody characterization as needed for the parallel determination of binding parameters and the detection of undesired cross-reactivities.
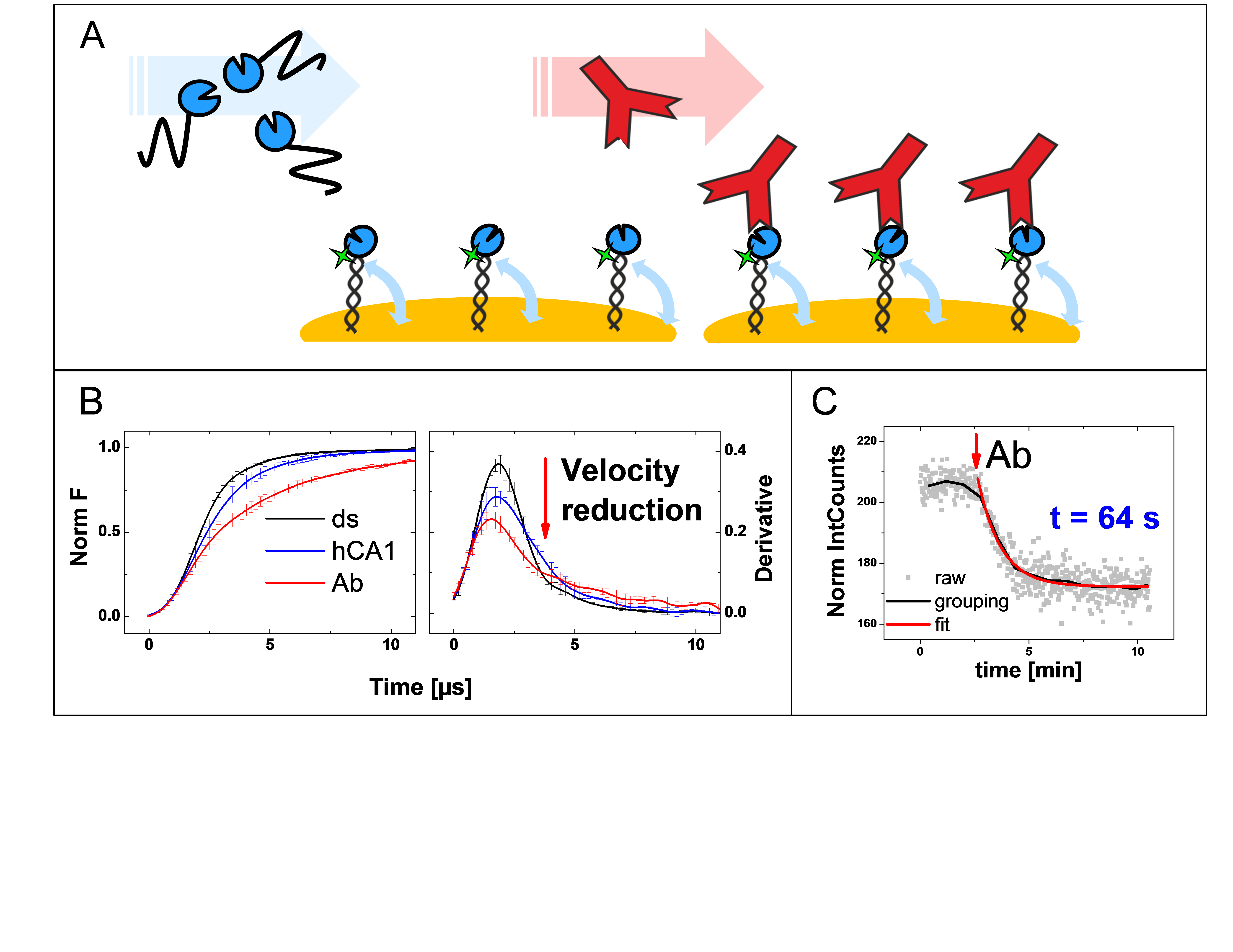
- Fig. 1: A - Schematic experimental method of antibody/antigen affinity binding detection on electrically actuated DNA layers, in a label-free manner. B – Molecular dynamics of the electrically modulated layers is used to identify the binding state of molecules, due to the increase in hydrodynamic drag, which slows the switching movement; (left) normalized fluorescence signal with error bar of standing motion and (right) its first derivative for in black initial ds-DNA, in blue hybridized hCA1-DNA conjugates, in red detection of bound antibody. C – Real time monitoring of antibody binding on hCA1 functionalized layers; 10 nM of antibody is loaded and the affinity can be estimated in molecular dynamics fitted data, in red (t time constant equal to 64 s in this case).
Institute of Photonic Technology, Albert-Einstein-Straße 9, 07745 Jena, Germany
janina.wirth@ipht-jena.de
Upon electromagnetic wave excitation, metal nanoparticles offer a high absorption and/or scattering, if the frequency of the electromagnetic wave is in resonance with the electron oscillation frequency. This collective oscillation of the electrons is termed Localized Surface Plasmon Resonance (LSPR) and depends on different properties of the nanoparticle, e.g. its material and size.
The high light absorption by the nanoparticles in combination with short laser pulses enables a contact-free manipulation of biological matrices, such as cells and DNA molecules. Hereby, the interaction between the laser pulses and the nanoparticles plays an important role. While the excitation of metallic nanoparticles by nano- or picosecond laser pulses results in a temperature increase in the dielectric surrounding, the excitation by femtosecond pulses can cause the escape of electrons without any temperature increase in the particle environment [1]. Thus, the pulse width of the laser has a significant impact on how the nanoparticle environment is affected.
Here, we present our studies about the interaction of nano- and femtosecond laser pulses with silver (AgNPs) and gold nanoparticles (AuNPs) in dependence of the laser fluence as well as their application in cancer cell treatment and DNA manipulation [2]. While the exposure of AuNPs labeled cancer cells with ns-laser pulses affords destruction at single cell level (Fig.1., right image), destruction of AgNP labeled DNA molecules by fs-laser pulses takes place in the nanometer length scale (Fig.2, right image) [3, 4].
- Fig. 1: ns-laser exposure of AuNPs (left) and destruction of AuNP-labeled cancer cells at a fluence of about 150mJ/cm2 and a wavelength of 530nm.
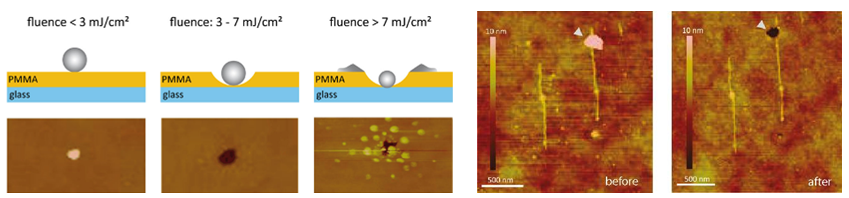
- Fig. 2: Effects of different laser fluences on AgNPs (left) and manipulation of AgNP-labeled DNA molecules (right) by fs laser exposure at a certain fluence of about 3mJ/cm2 and a wavelength of 800nm.
1] Garwe, F., et al., Nanotechnology, 2008. 19(5).
[2] Csaki, A., et al., Nano Letters, 2007. 7(2): p. 247-253.
[3] Csaki, A., et al., Philosophical Transactions of the Royal Society a-Mathematical Physical and Engineering Sciences, 2011. 369(1950): p. 3483-3496.
[4] Wirth, J., et al., Nano Letters, 2011. 11(4): p. 1505-1511.
1. Lab of Biophotonics & Biosensing, Max Plank Institute for the Science of Light
2. Wyss Institute for Biologically Inspired Engineering, Harvard University
yuqiang.wu@mpl.mpg.de
Whispering gallery mode resonators have been proved to be highly sensitive for label-free DNA detection [1]. DNA toehold exchange technique opens a door to precisely control DNA strand displacement [2]. By combining these two techniques, we design three mechanisms: translator, aggregation and catalyst, which enable us to detect arbitrary but known sequences of DNA, increase the signal to noise ratio of the sensor, and achieve high sensitivity. Moreover, for detecting different targets, we use the same type of surface-modified microspheres, simply with different programed DNA 'translators'.
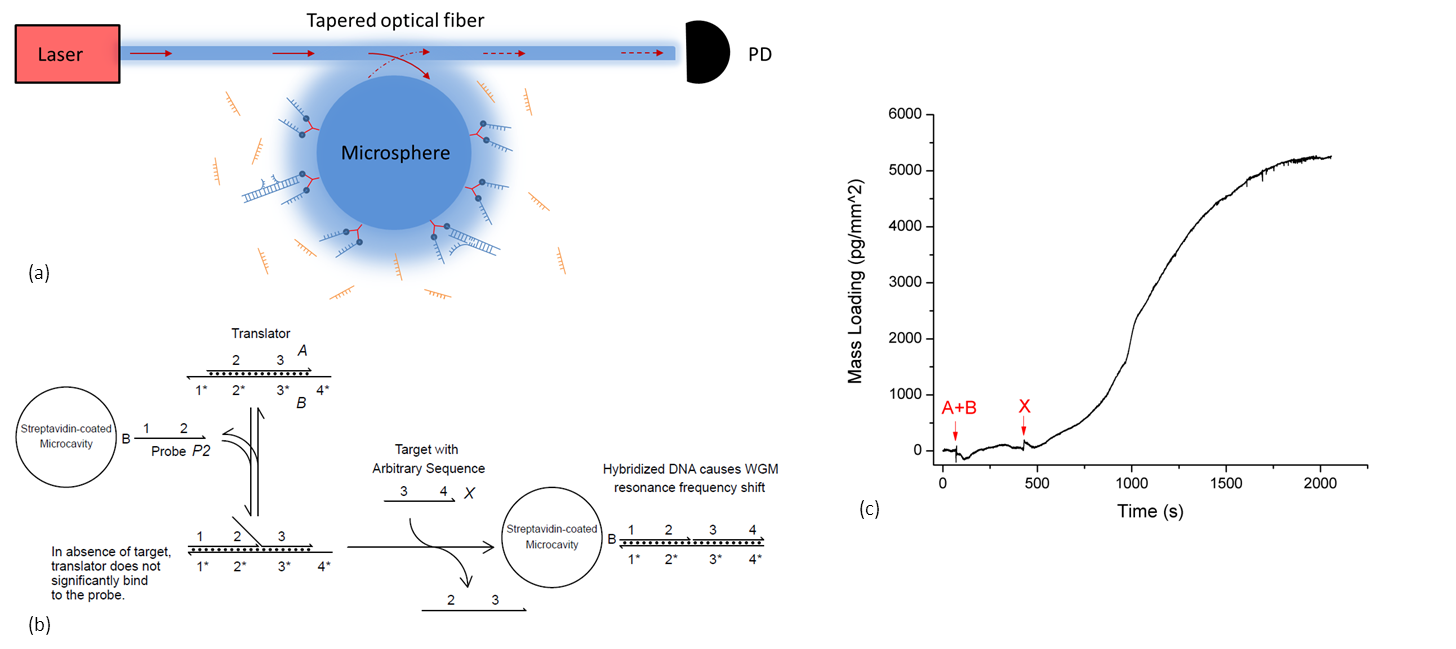
- Fig. 1: (a)- Schematic of the experimental setup. (b) – The 'translator' mechanism based on toehold exchange technique. (c) – Experimental results showing the detection of arbitrary sequence X; concentration for all sequences is 500 nM.
[1] Vollmer F., Arnold S., Braun D., Teraoka I., Libchaber A., Multiplexed DNA Quantification by Spectroscopic Shift of Two Microsphere Cavities, Biophysical Journal, 3, 85: 1974-9 (2003).
[2] David Y Zhang and Erik Winfree, Control of DNA Strand Displacement Kinetics Using Toehold Exchange, J. Am. Chem. Soc. 131: 17303-17314 (2009).