
1 - Center for DNA Nanotechnology, University of Aarhus, Langelandsgade 140, 8000 Århus C
2 - Department of Chemistry and the Biodesign Institute, Arizona State University, Tempe, Az, 85287 (USA)
The poster is describing how covalently linked organic nanostructures have been prepared using DNAdirected synthesis. Furthermore, the poster describes recent results achieved in forming organic wires using a DNA template or a DNA nanostructure to control the reaction. The synthesis involved in the formation of the large organic − DNA conjugates will be shown.
Division of Pathway Medicine, University of Edinburgh Medical School
The Chancellor's Building, 49 Little France Crescent, Edinburgh, EH16 4SB, UK
till.bachmann@ed.ac.uk
Infectious diseases are one of leading causes of human illness and death. More than 16 million fatalities every year can be accounted to an infection by bacteria, viruses, parasites, or fungi. Examples of the most threatening pathogens are well known cases like the Hepatitis C virus but also those causing emerging infectious diseases such as SARS. Other, mostly less life threatening infections, for example urinary tract infections (UTI), cause still a severe damage to human well-being and the society's economy. In addition, over the last decades antimicrobial resistance became a severe issue and a threat the general public became increasingly aware. Therapy of microbial infections requires ideally a profound diagnostics comprising rapid detection of the causing agent's identity and virulence to enable the choice of the right chemotherapy. Conventional methods are mostly too laborious, costly, slow and do not allow near patient testing to the desired extent. Associated with these facts, antimicrobial agents are sometimes misused fuelling antimicrobial resistance development. In contrast, biochips enable rapid detection and genotyping of pathogens and can be integrated in microfluidic settings providing a rapid time to result and point of care testing. Furthermore, biochips allow the integration of pathogen identification with therapeutic drug monitoring as well as the host response to infection. Such approaches combine drug therapy and diagnostics to theranostics supporting personalised healthcare. The presentation covers our recent developments on fluorescent DNA chips for pathogen identification (e.g. sepsis, gastroenteritis, Hepatitis C, UTI), virulence (e.g. pathoadaptive mutations), and antibiotic resistance (e.g. Quinolone resistance, Extended Spectrum Beta Lactamases) and clinical validation data. The presentation will finally detail our recent technical progress on advanced detection technologies using DNA nanoswitches, e.g. on SNP detection.
[1] Barl,T, Dobrindt,U, Katcoff,D, Bingen,E, Hacker,J, Bachmann,TT; Genotyping pathoadaptive mutations in the type 1 fimbriae,
submitted
[2] Barl,T, Dobrindt,U, Yu,X, Katcoff,D, Bingen,E, Hacker,J, Bachmann,TT; Genotyping DNA chip for the simultaneous assessment of
antibiotic resistance and pathogenicity potential of extraintestinal pathogenic Escherichia coli, submitted
[3] Weile J, Susa M, Schmid RD, Bachmann TT, Knabbe C; DNA-microarray for genotyping multidrug resistant P. aeruginosa clinical
isolates, Diagn Microbiol Infect Dis, in print
[4] Yu X, Susa M, Weile J, Knabbe C, Schmid RD, Bachmann TT; Rapid and sensitive detection of fluoroquinolone-resistant
Escherichia coli from urine samples using a genotyping DNA microarray. Int J Med Microbiol. 2007 May 4
[4] Strommenger B, Schmidt C, Werner G, Roessle-Lorch B, Bachmann TT, Witte W; DNA microarray for the detection of
therapeutically relevant antibiotic resistance determinants in clinical isolates of Staphylococcus aureus. Mol Cell Probes. 2006 Oct
19
[5] Leinberger DM, Schumacher U, Autenrieth IB, Bachmann TT; Development of a DNA microarray for detection and identification of
fungal pathogens involved in invasive mycoses. J Clin Microbiol. 2005 Oct;43(10):4943-53
[6] Grimm V, Ezaki S, Susa M, Knabbe M, Schmid RD, Bachmann TT; Rapid resistance genotyping of TEM beta-lactamases using
DNA-microarrays, J Clin Microbiol, 2004, 42: 3766-3774
[7] Buck AH, Campbell CJ, Dickinson P, Mountford CP, Stoquert HC, Terry JG, Evans SA, Keane LM, Su TJ, Mount AR, Walton AJ,
Beattie JS, Crain J, Ghazal P. DNA nanoswitch as a biosensor. Anal Chem. 2007 Jun 15;79(12):4724-8.
[8] Mountford CP, Buck AH, Campbell CJ, Dickinson P, Ferapontova EE, Terry JG, Beattie JS, Walton AJ, Ghazal P, Mount AR, Crain
J. Molecular recognition with DNA nanoswitches: effects of single base mutations on structure. J Phys Chem B. 2008 Feb
28;112(8):2439-44.
Fraunhofer Institute for Biomedical Engineering, Am Mühlenberg 13, 14476 Potsdam-Golm, Germany frank.bier@ibmt.fraunhofer.de
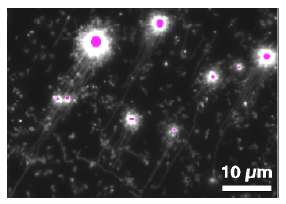
Fluorescence micrograph of ssDNA strands having being synthesised on
nanobeads (purple) and hydrodynamically stretched and aligned
For a controlled interaction of DNA based nanodevices with the macroscopic world a well defined contact to surfaces is advisable. For this purpose we have developed two complementary methods:
DNA based self-organisation allows the assembly of nanodevices into larger structures, since distinct positions on a DNA-scaffold can be specifically addressed by their base sequence. For this we have developed a system for the surface anchored synthesis of single stranded DNA by rolling circle amplification. Being integrated into a flow-through system with optical access, the DNA molecules can be fluorescently stained, hydrodynamically stretched and then hybridised with complementary nucleotides that are tagged to nano-objects. Surface properties have been adjusted to control the adhesion of both educts and products. Parallel, stretched ssDNA molecules of several tens of micrometres have been synthesised.
For the second method the tip of a scanning force microscope is used to "write" oligonucleotides onto surfaces by molecular ink lithography with great positional accuracy. Coupling to the surface is accomplished with methods adapted from microarray technology, in this case covalent coupling of amino-modified oligonucleotides to epoxy-functionalised glass surfaces. Since the supply of molecules from the scanning tip to the surface is mediated by a water meniscus, temperature and relative humidity are controlled by placing the setup into a climatised chamber. Loading of the tip is performed through a fluidic system. The system currently allows the routine preparation of nanoarrays with 3x3 spots of 300 nm size at arbitrary positions.
Department of Biochemistry, George S. Wise Faculty of Life Sciences, Tel Aviv University, Ramat Aviv, 69978 Israel
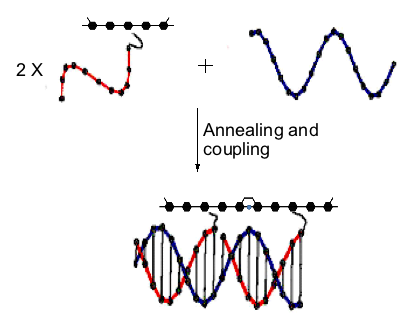
G-tetrad is a unique structural arrangement of four guanine bases, specific to guanine-rich DNA sequences. It is a fundamental binding block of the higher order nucleic acid structure known as a G-quadruplex or G4- DNA. Because of its high content of guanines, which have the lowest ionization potential among DNA bases and remarkable structural features, G4-DNA molecule has attracted a special interest in the field of nanotechnology as a potential conductor of electrical current. So far, the only long continuous G4-DNA reported nanostructures are monomolecular quadruplexes (Kotlyar, A. B., Borovok, N., Molotsky, T., Cohen, H., Shapir, E., Porath, D. 2005, Adv. Mater. 17, 15, 1901-1905), made of a single G-strand. Long Gquadruplexes composed of four long parallel G-strands is another intriguing structure that might possess improved electric properties with respect to the monomolecular G4-DNA explored so far. However, intermolecular G4-wires do not form spontaneously in solutions. At high concentration, which is required for the tetra-molecular assembly, G-strands aggregate into bundles comprising of a large number of strands. To overcome these problems and to promote a tetra-molecular G4-DNA assembly, we developed a novel approach based on the avidin-biotin recognition. By using avidin, a tetramer glycoprotein, it is possible to bring together four G-strands, end-labeled with biotin, and enable their integration into a parallel tetramolecular structure. The overall scheme of the synthesis is shown below.
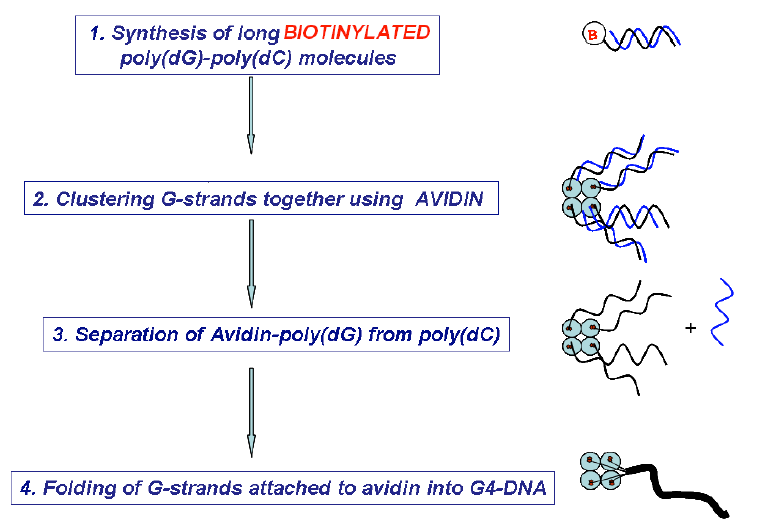
This method enabled to prepare uniform continuous G4-DNA nanostructures composed of four parallel Gstrands. The results of AFM imaging of the molecules showed that their average contour length is approximately equal to that of a parent poly(dG)-poly(dC) proving the tetramolecular mechanism of the Gstrands folding into the G4-quadruplex.
Fraunhofer Institute for Biomedical Engineering, Am Mühlenberg 13, 14476 Potsdam-Golm, Germany
ralph.hoelzel@ibmt.fraunhofer.de
Nanoscale constructs based on nucleic acids can be prepared exploiting self-assembly processes. To apply these constructs they usually have to be connected to the macroscopic world. This contact to a surface can be accomplished with the help of an atomic force microscope (AFM). For this the AFM tip is loaded with a reaction partner which is released upon contact to the surface ("dip pen nanolithography"). This phenomenon can be explained by diffusion through a water meniscus that is built up between surface and tip.
In by far most cases a gold-to-thiol reaction is used for coupling to the surface. Here we present results exploiting covalent coupling between an epoxy-modified glass surface and amino-modified oligonucleotides. This should pave the way for downscaling methods that have already been well established in micro-array technology for the production of nano-arrays.
The supply of molecules from the AFM tip to the surface strongly depends on the water meniscus' properties and, hence, on temperature and relative humidity. Therefore we have placed the complete AFM head into a climatised chamber. A fluid system was designed that allows loading of the tip with different substances and that still gives good reproducibility concerning tip position. The influence of physical parameters as well as of additives to the DNA solution was tested.
Currently nano-arrays with 3x3 spots of 300 nm diameter each are routinely prepared. Micro-arrays of more than 100 spots with 2 μm size can be produced with slight modifications of the protocol. The method will be applied to the analysis of minute volumes, e.g. of single cells, as well as to the anchoring of nanodevices.
Laboratoire d'Electronique Moléculaire CEA Saclay, DSM/DRECAM/SPEC, 91191 Gif/Yvette, France Stephane.Campidelli@cea.fr
In nanotechnology self-assembly is a valuable strategy where the design, manufacture and control on the scale of a few nanometers are governed by molecular and supramolecular affinities (like structural and chemical properties). In particular, the exceptional recognition properties of DNA make it an ideal candidate to encode instructions for such nano-scale assembly to create scaffolds [1, 2] and to incorporate other materials in the self-assembly process [3]. The use of DNA not only as a positioning scaffold but also for electrical interconnections is highly desirable in a nanoelectronic context. Since DNA transport properties are still under discussion [4], it is pragmatically envisioned to develop a method where electronic conduction is ensured by a metallic coating selectively deposited onto the DNA strand.
In our approach we decided to focus onto Pd metalization since it has been shown to be the best choice for electrodes connecting single wall carbon nanotubes (SWNTs). SWNTs occupy a special place within molecular electronics [5] and we give a particular attention is given to methods able to self assemble and electrical connect these nanoobjects. In the DNA-directed vision the metallization process should be the last step of the fabrication process to keep as long as possible DNA recognition properties. Therefore we performed our study on DNA deposited onto the substrate to obtain metallic nanowires while minimizing parasitic metal cluster nucleation and growth on the surrounding surface. In addition, our concern in this study is related to both yield uniformity and conductive behaviour of the so obtained nanowires. These two issues related to the metallization process are really crucial for the use of DNA scaffolding in a circuit context where neither a big resistance value nor a large desorption are acceptable.
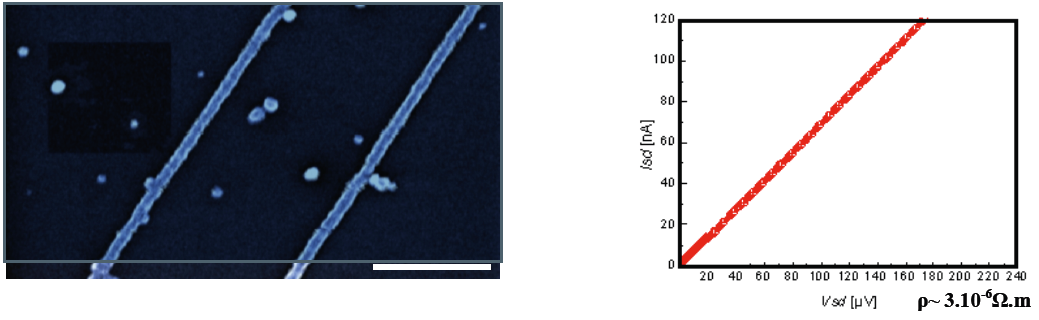
Figure 1: Conductive Pd nanowires synthesized on DNA scaffolds. Left hand side: scanning electron microscope image (estimated diameter ~ 25 nm). Right hand side: I/V characteristic showing conductive behaviour
This work has been partially supported by the NUCAN - NMP STREP 013775 project
[1] N. C. Seeman, Nature 421 (2003), 33-37
[2] N. C. Seeman, Nanotechnology 2 (1991), 149-159
[3] C. A. Mirkin, Inorg. Chem. 39 (2000), 2258-2272
[4] X Guo et al., Nature Nanotechnology 3 (2008), 163-167. Xu et al, Nanoletter 4(2004)1105-1108 . Storm et al. App.Phys.Lett. 79
(2001) 3881-3883
[5] Avouris et al, Nature Nanotechnology 2 (2007), 605-615
Laboratoire d'Electronique Moléculaire, DSM/IRAMIS/SPEC,
CEA Saclay, 91191 Gif sur Yvette, France.
Email: chia-ling.chung@cea.fr
Nanometer-scale structures represent a novel and intriguing field, where scientists and engineers manipulate materials at the atomic and molecular scale levels to produce innovative materials. Carbon nanotubes constitute a relatively new class of materials exhibiting exceptional mechanical and electronic properties and were found to be promising candidates for molecular electronics, sensing or biomedical applications.1 Considering the bottom-up strategy in nanotechnology, the combination of the recognition properties of DNA with the electronic properties of single walled carbon nanotubes (SWNTs) seems to be a very promising approach for the future of electronics.2
With the aim to assemble DNA with SWNTs, two complementary strategies have been envisioned: the covalent linkage of DNA on carboxylic groups of SWNTs under classical carbodiimide coupling condition3 and the non-covalent approach based on biotin-streptavidin molecular recognition properties.4 Here, we present and compare the results that we obtained with these two different methods; we want to objectively show the advantages and disadvantages of each approach.

Figure. AFM images of covalently linked DNA-SWNT (left) and non-covalent DNA-Streptavidin-SWNT assemblies (right); in both images the scale bars represent 200 nm.
This work has been partially supported by the NUCAN − NMP STREP 013775 project and CHEMTRONICS MEST-CT-2005-020513
[1] a) E. Katz, I. Willner, ChemPhysChem, 2004, 5, 1084. b) M. J. O'Connell, Carbon Nanotubes: Properties and Applicatons; CRC
Press: 2006.
[2] a) K. Keren, R. S. Berman, E. Buchstab, U. Sivan, E. Braun, Science, 2003, 302, 1380. b) K. Nguyen et al., in preparation.
[3] a) C. Dwyer, M. Guthold, M. Falvo, S. Washburn, R. Superfine, D. Erie, Nanotechnology, 2002, 13, 601. b) M. Hazani, R. Naaman,
F. Hennrich, M. M. Kappes, Nano Lett., 2003, 3, 153.
[4] S. Lyonnais, L. Goux-Capes, C. Escudé, D. Cote, A. Filoramo, J.-P. Bourgoin, Small, DOI: 10.1002/Smll.200700586.
1 Friedrich-Schiller-University Jena, Department of Physical Chemistry, Jenaer Biochip Initiative (JBCI), Helmholtzweg 4, 07743 Jena, Germany
2 Institute of Photonic Technology Jena (IPHT), Albert-Einstein-Straße. 9, 07745 Jena, Germany
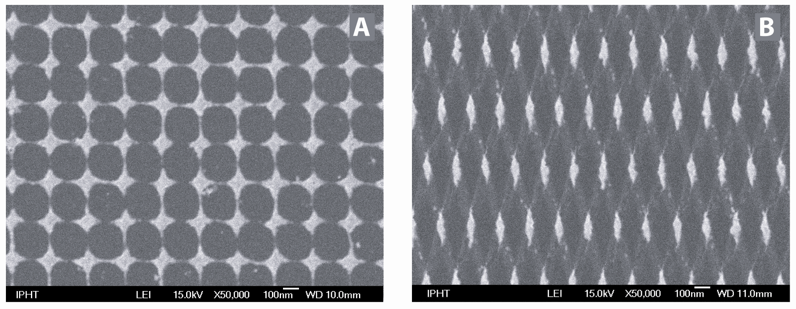
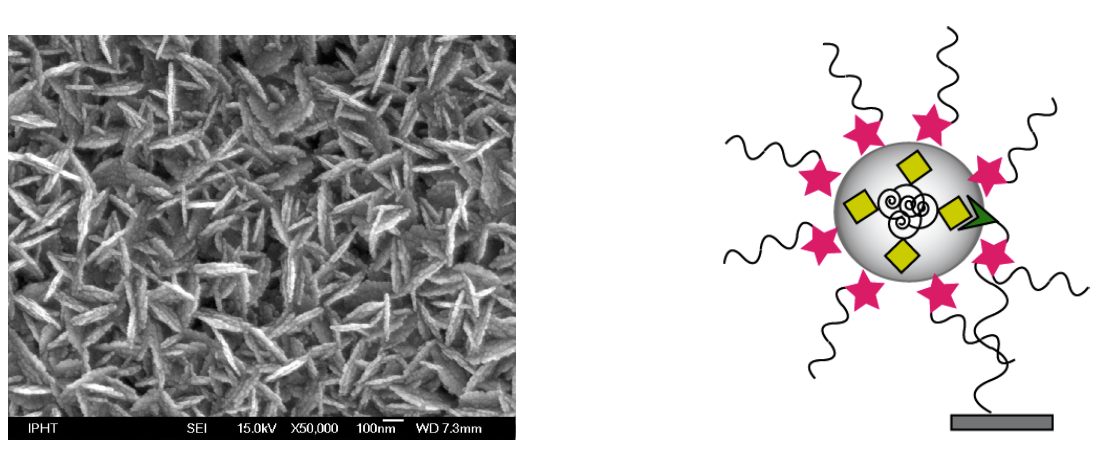
Within this contribution new types of microfabricated SERS active substrates produced by electron beam lithography and ion beam etching are introduced. In order to achieve large enhancement factors by using the lightning rod effect we prepared arrays consisting of sharp edged nanostructures instead of dots, which are commonly used. [1, 2] Two experimental methods were used for fabrication: a one-stage process, leading to gold nanostar arrays and a two-stage process, leading to gold nanodiamond arrays. Our preparation process guarantees a high reproducibility. For practical application, the substrates contain a number of arrays, each 200 x 200 μm2 in size. To test the SERS activity of these nanostar and nanodiamond arrays a monolayer of the dye crystal violet was used. Enhancement factors were estimated to be at least 130 for the nanodiamond and 310 for the nanostar arrays, respectively. [3]
In further research the Raman excitation wavelength providing best possible enhancement will be determined by recording UV-Vis absorption spectra using a modified spectrophotometer with a parallel beam path less than 200 μm in diameter. The substrates will also be tested using a non-resonant analyte (without molecular resonance nearby the excitation wavelength) as preparation for an applicative DNA detection. A dye labeled as well as label free DNA detection scheme using the microfabricated SERS substrates reported within this contribution and also newly fabricated ones is currently under way.
Acknowledgement
The research project "Jenaer Biochip Initiative (JBCI)" within the framework "InnoProfile − Unternehmen Region" is financially supported by the Federal Ministry of Education and Research (BMBF) Germany.
[1] L. Billot, M. Lamy de la Chapelle, A. S. Grimault, A. Vial, D. Barchiesi, J. L. Bijeon, P. M. Adam, P. Royer, Chemical Physics Letters
2006, 422(4-6), 303-307.
[2] M. Sackmann, S. Bom, T. Balster, A. Materny, Journal of Raman Spectroscopy 2007, 38(3), 277-282.
[3] D. Cialla, U. Hübner, H. Schneidewind, R. Möller, J. Popp, ChemPhysChem 2008, accepted for publication.
Institute of Photonic Technology (IPHT), Jena, A.-Einstein-Str. 9, 07745 Jena
*Fraunhofer Institut für Zuverlässigkeit und Mikrointegration (IZM), Berlin, Dept. Module Integration and Board Interconnection Technologies, Gustav-Meyer-Allee 25, 13355 Berlin
Dielectrophoresis describes the behavior of neutral bodies in AC-field in dependence on their polarizability [1]. Using this method an accumulation [2], separation and directed electrocasting of particles [3] or integrating of biomolecules [4] are possible. In our case dielectrophoresis will be used for the contact-free generating of metal nanowires between microelectrodes. So the contacting of bottom-up nanoscale objects to top-down technical periphery like microelectrodes from classical photolithographic process is possible. Investigations of parameters and key factors for the optimum building of nanowires from metal nanoparticles and metallized vesicles were performed. Particle types, concentrations, used frequencies, voltages and reaction times for each layout type were summarized in parameter complexes. It could be demonstrated that the resulting defined nanowires are appropriated for nanoelectronic and sensoric applications.
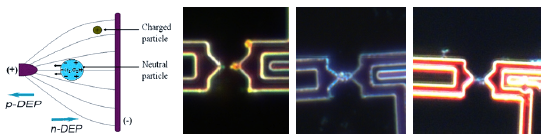
Principle of dielectrophoretis (left). Field intensity spikes visualized with 60 nm gold nanoparticles (center left). Nanoparticle nanowires with 60 nm gold particles and metallized vesicles (right).
[1] Pohl, H. A. and I. Hawk (1966). "Separation of Living and Dead Cells by Dielectrophoresis." Science 152(3722): 647-649.
[2] Holzel, R. (2002). "Single particle characterization and manipulation by opposite field dielectrophoresis." Journal of Electrostatics
56(4): 435-447.
[3] Fiedler, S.; Zwanzig, M.; Schmidt, R.; Auerswald, E.; Klein, M.; Scheel,W.; Reichl, H. Evaluation of Metallic Nano-Lawn Structures
for Application in Microelectronic Packaging. IEEE 1st Electronics Systemintegration Technology Conference 2006, September 5th-
7th, Dresden, Germany. Proc. (2006) pp. 886-89
[4] Wolff, A.; Ch. Leiterer; A. Csáki; W. Fritzsche, Dielectrophoretic manipulation of DNA in microelectrode gaps for single-molecule
constructs, Frontiers in Bioscience (2008), in press
Institute of Photonic Technology (IPHT), Jena, A.-Einstein-Str. 9, 07745 Jena
DNA-based nanotechnology offers a tremendous potential for the construction and integration of defined nanoscale construct via self-assembly. These mechanical stable, flexible molecules can build a template for nanoscale structures while also being appropriate for the biological functionalization of different surfaces. This biological matrix has a highly defined structure and its sequence ensures a direct addressability for manipulation on the nanoscale level.
Defined constructions from different metal nanoparticles (gold, silver and bimetallic core-shell) were demonstrated using DNA-DNA interaction. Additionally, we compared immobilization techniques for framework molecules enabling a technological implementation in a parallel way. Basic criteria for the immobilization are the addressability, specificity and the defined geometry of the immobilized molecules. The addressability enables the site-directed assembly of framework molecules in parallel. The specificity ensures their selective binding onto surfaces whereas a defined geometry, i.e. a geometrically appropriate arrangement, is required for the following manipulation steps in the nanoconstruction. We therefore tested combined stretching and immobilization methods for framework DNA molecules and DNA superstructures like G-wires. The one- or two-step binding of these molecules was arranged on microelectrodes from conventional photolithographic techniques. The applied techniques, like immobilization and stretching by hydrodynamic and electrical forces, were tested for their effectivity and ability for the construction of future nanoelectronic devices. As last step of the construction, specific metallization of DNA molecules was used for the generation of small nanowires enabling the electrical contacting to the technical periphery (electrodes). On one hand, we realized this metallization by binding of gold nanoparticles as seeds for a subsequent silver deposition. On the other hand, we arranged this by direct metallization along the DNA backbone.
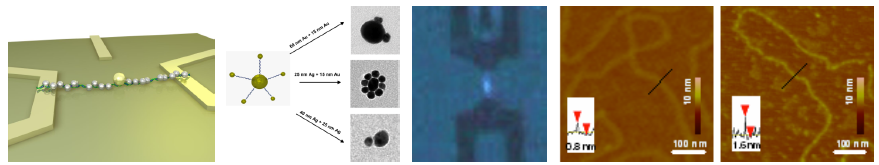
Principle of DNA-based SET devices (left). Nanoscale particle constructs using DNA self-assembly (center left). Microintegration of framework molecules via receding meniscus and metallization of the molecules (right).
[1] Csáki, A., G. Maubach,... (2002). "DNA-based molecular nanotechnology." Single Mol. 3(5-6): 275-280.
[2] Maubach, G. and W. Fritzsche (2004). "Precise Positioning of Individual DNA Structures in Electrode Gaps by Self-Organization
onto Guiding Microstructures." Nano Letters 4(4): 607-611.
[3] Maubach, G., D. Born,... (2005). "Parallel Fabrication of DNA-Aligned Metal Nanostructures in Microelectrode Gaps by a Self-
Organization Process." Small 6: 619-624.
[4] Marsh, T. C., J. Vesenka, et al. (1995). "A new DNA nanostructure, the G-wire, imaged by scanning probe microscopy." Nucleic
Acids Res 23(4): 696-700.
[5] Wolff, A.; Ch. Leiterer; A. Csáki; W. Fritzsche, Dielectrophoretic manipulation of DNA in microelectrode gaps for single-molecule
constructs, Frontiers in Bioscience (2008), in press
1: University of Oxford, Department of Physics, Clarendon Laboratory, Parks Road, Oxford, OX1 3PU, United Kingdom
2: Present address: Castle Society, Harvard Medical School, 260 Longwood Avenue, Boston, MA, 02215, USA
c.erben@physics.ox.ac.uk

Figure 1: A single molecule of cytochrome c inside a DNA tetrahedron (C. M. Erben, R. P. Goodman, A. J. Turberfield, Angew. Chem. Int. Ed. 45, 7414 (2006)).
Nanometre-sized polyhedra can be constructed from synthetic DNA oligonucleotides in a facile single-step self assembly process. The edges of these polyhedra are rigid DNA double helices joined by three- or fourarm junctions at the vertices.
A variety of different polyhedra have been formed, including tetrahedra, octahedra and trigonal bipyramids. We can control the topology of the resulting polyhedra by sequence design: DNA strands can be guided along the structure in different ways to create or avoid crossovers of strands at the vertices.
DNA polyhedra can be used as rigid three-dimensional scaffolds or as cages for other molecules. We have demonstrated the encapsulation of a single protein molecule inside a DNA tetrahedron (see Figure 1). Molecules can be attached to the DNA structure in different ways: we have explored new non-covalent linkage methods for the attachment of proteins to DNA.
DNA devices which are capable of controlled motion have also been constructed. Well-defined changes in their three-dimensional conformation can be triggered by the addition of DNA fuel strands and powered by the energy released by DNA hybridisation. We have shown that a reconfigurable DNA tetrahedron can be cycled between open and closed states in this way. Combination of encapsulation and triggered conformational changes suggests potential applications in drug delivery.
Laboratoire d'Electronique Moléculaire, CEA Saclay, DSM/IRAMIS/SPEC, 91191 Gif-sur-Yvette, France
Thanks to their exceptional electrical, mechanical and chemical characteristics, carbon nanotubes are very promising building blocks for future nanoelectronic technologies. However, the future of this class of SWNTbased devices is to a large extent related to the development of a bottom-up self-assembly technique. Indeed, at this scale self-assembly, and more generally, "bottom-up" approaches appear to be a more reasonable way to assemble nano-objects into circuits with a two-dimensional and/or three-dimensional layout.
Here, a bio-inspired DNA-directed approach will be presented: construction of the DNA scaffold, DNAnanotube chemistry, DNA metallisation and devices. This bio-directed method constitutes a genuine and complete molecular-scale bottom-up method, since it relies on recognition properties inherent to biological entities and can be employed at nanoscale without using any standard lithography technique.
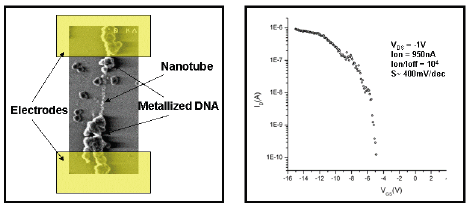
Figure 1: DNA-Carbon nanotube device. In the left hand side it is shown a SEM image of a device realised by DNA-carbon nanotube assembly. The DNA has been selectively metalized. In this picture the lithographically defined electrodes have been schematized. In the right hand side the I/V curve of such device is reported showing transistor characteristic.
This work has been partially supported by the NUCAN − NMP STREP 013775 project
Technische Universität München, Germany
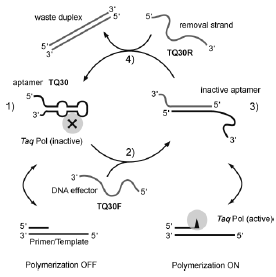
The molecular recognition properties of DNA can be utilized to construct nanomechanical devices that can be reversibly switched between distinct conformations. Functionality of such a devices can be achieved by including aptamer motifs into such devices. Aptamers are short stretches of singlestranded DNA selected from a random pool of DNA sequences for their binding affinities to proteins or small molecules.
By this, we constructed a molecular machine based on a DNA aptamer, which can be instructed to "grab" or release the human blood-clotting factor, - thrombin, depending on an operator DNA sequence addressing it. In the operation of this DNA nanomachine, the thrombin-binding DNA aptamer is mechanically switched between a binding and a nonbinding form: The known sequence for a thrombin-binding aptamer was modified with a random 12-nucleotide (nt) sequence - a toehold - at which a DNA strand partly complementary to the aptamer sequence could attach. This effector or fuel strand could then displace the protein from the aptamer by binding to the aptamer sequence. A second toehold on the effector strand was used to remove the effector strand from the aptamer by a removal strand. In this process, the aptamer's binding capabilities were restored and the protein was bound again [1].
We could demonstrate the general use of this approach -utilizing operator DNA strands to control the device - by applying it to a second system, a DNA aptamer machine capable of binding to DNA polymerase from Thermus aquaticus (Taq polymerase). The result is a simple molecular device that allows us to control the enzymatic activity of Taq polymerase. In contrast to ref. [1], we show here that a switchable aptamer can actually be used to switch the biological activity of an enzyme reversibly [2].
[1] Dittmer, WU; Reuter, A; Simmel, F.C. Angewandte Chemie International Edition 2004, 43, 27, 3550.
[2] Friedrichs, E; Simmel, F.C., Chembiochem 2007, 8, 1662.
1 Center for Functional Nanomaterials, 2 Biology Department, Brookhaven National Laboratory, Upton, NY 11973, USA ogang@bnl.gov
Incorporation of DNA into nano-object design provides a unique opportunity to establish highly selective and reversible interactions between the components of nanosystems. Utilization of this approach is appealing for development of novel strategies for the assembly of complex nanosystems, metamaterials and nano-medical applications. The presented work explores how DNA can be engaged in addressable interactions of inorganic nano-components, and how kinetics and morphology of self-organized structures can be regulated. For example, by tailoring the design of DNA shell we have demonstrated an ability to control assembly kinetics [5], clustering [3,4], and phase formation of DNA-capped nanoparticles on surfaces and in bulk [1,2]. Using synchrotron small-angle x-ray scattering we recently observed formation of 3D crystalline structures with body centered cubic order in two-component nanoparticle systems, where DNA functionalized particles either hybridize directly [1] or via DNA linkers [2]. The design principles and experimental illustrations of meso- and nano- hybrid systems will be discussed.
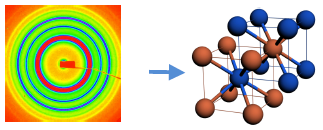
Figure. Small-angle x-ray scattering image (left) and illustration of the corresponding bcc lattice (right), where blue and red colors represent particles with complementary DNA [1].
[1] D. Nykypanchuk, M. M. Maye, D. van der Lelie, and O. Gang, "DNA-guided crystallization of colloidal nanoparticles", Nature 451,
549-552 (2008)
[2] H. Xiong, D. van der Lelie, and O. Gang, "DNA Linker-Mediated Crystallization of Nanocolloids", J. Am. Chem. Soc, 130 (8), 2442
(2008)
[3] M. M. Maye, D. Nykypanchuk, D. van der Lelie, and O. Gang, "DNA-Regulated micro- and nanoparticle assembly", Small 3, 1678
(2007)
[4] D. Nykypanchuk, M.M. Maye, D. van der Lelie, Daniel, and O. Gang, "DNA-based approach for interparticle interaction control",
Langmuir 23 (11): 6305-6314, (2007)
[5] M. M. Maye, D. Nykypanchuk, D. van der Lelie, and Oleg Gang "A Simple Method for Kinetic Control of DNA-Induced Nanoparticle
Assembly", J. Am. Chem. Soc., 128 (43), 14020 (2006)
1 Laboratoire d'Electronique Moléculaire, CEA Saclay, France.
2 Laboratoire de Chimie de la Reconnaissance et Etude des Assemblages Biologiques, CEA Grenoble, France
Progress in micro-electronic, surface chemistry, biology and computing has allowed a lot of useful applications for DNA modified surfaces, such as mutation analysis, genes expression analysis, sequencing and medical diagnosis. Grafted DNA on substrates can also constitute a nano-scale platform allowing the immobilization of little objects [1]. This approach provides an interesting way to study cascade reactions between proteins confined in a restraint space, designed specifically for the studied reaction [2]. The aim of this work is to use a DNA strand, grafted on gold electrodes, as a platform to immobilize nanoobjects (metallic nanoparticules, proteins...).
Single stranded oligonucleotides functionalized surfaces are required for the anchorage of the DNA of interest. Among all the functionalization strategies employed for the development of sequence-specific tools, we have chosen to synthesize DNA-polypyrrole films [3]. An electro-copolymerisation of pyrrole units modified with a single stranded DNA and non-modified pyrrole units provides the grafting of the probes suitable for the immobilization of a DNA strand on our gold micro-electrodes. A DNA target, modified in one part by biotin entities, is immobilized by hybridization with the probes already fixed. Assemblies between grafted DNA and streptavidin modified entities can be perfomed thanks to the affinity biotin-streptavidin.
This work has been partially supported by the French government via Bio-NT project and by European commission NUCAN − NMP STREP 013775 project
[1] T. H. LaBean, H. Li, Nano Today, 2007, 2, 26.
[2] C. M. Niemeyer, Nano Today, 2007, 2, 42.
[3] T. Livache, B. Fouque, A. Roget, J. Marchand, G. Bidan, R. Téoule, G. Mathis, Analytical Biochemistry, 1998, 255, 188.
Nanoprobes, Incorporated, 95 Horseblock Road, Unit 1, Yaphank, NY 11980, USA
Horse radish peroxidase (HRP) is a commonly used enzyme for detection in applications such as ELISAs and immunohistology. Typically, substrates producing a color are used, such as diaminobenzadine (DAB) which gives an insoluble brown color, or supplemented with nickel or cobalt to give a gray-black color, 3,3',5,5'-tetramethylbenzidine (TMB) to give an insoluble blue color, 3-amino-9-ethylcarbazole (AEC) to give a red color, 2, 2'-azino-di(3-ethylbenzthiazoline-6-sulfonate) (ABTS), to give a soluble blue-green color, or 4- chloro-1-naphthol to produce an insoluble blue-gray color. Higher sensitivity has been obtained by adding metal ions such as nickel to DAB, or using a chemiluminescent substrate. A complementary method is Tyramide Signal Amplification (TSR), which uses HRP to deposit hydroxylated compounds such as fluorescent tyramides or biotin tyramide at target sites. Multiple biotins are deposited, giving more targets for streptavidin reporters. Alternatively, polymers of HRP, which can amplify signal strength, have been used. Examples include antibody-HRP aggregates cross-linked with glutaraldehyde, or antibodies and HRP conjugated to a dextran backbone. HRP is preferred due to its robustness as an enzyme, and its straightforward conjugation to antibodies, peptides, nucleic acids or other targeting molecules.
Although the typical reaction mechanisms for HRP involve oxidation of an organic substrate or creation of free radicals, Nanoprobes has discovered how to use HRP to reduce metal ions from solution, thus depositing insoluble solid metal in the zero oxidation state. This was shown to be a highly sensitive and specific staining and detection method, and carries a number of advantages over organic substrates: a) the metal deposition is highly localized and does not diffuse as DAB does, enabling high resolution staining, even at the EM level; b) the metal is more dense than organic dyes, thus giving greater contrast and visibility; c) the sensitivity is as good as or better than the most sensitive of the other HRP substrates, including chemiluminesent ones; d) the deposited metal can be used for the formation of electrically conductive sensing elements, for example in chip arrays where the metal bridges electrodes producing an electrical signal, whereas other HRP substrates must be read by more complex and expensive optical systems; e) the metal deposits are permanent, and do not fade, a problem with fluorescent products; f) the reaction is rapid, and high sensitivity can be achieved in minutes compared to many hours required when using chemiluminescence.
Due to the many advantages of this system, a commercial breast cancer test using EnzMetTM has been introduced that detects single HER2 genes by in situ hybridization (similar to FISH, but bright field and permanent) to detect HER2 gene amplification in biopsy samples. This mutation occurs in about 30% of breast cancer patients: it is associated with aggressive malignant behavior and poor prognosis, and if it is found, it is an indicator for treatment with monoclonal antibody (Herceptin) therapy, which can provide dramatic therapeutic benefits to patients with an otherwise poor prognosis. A further advance has been to convert the metal product into a colored one such that multiple markers can be distinguished.
Chemical Nanoscience Laboratories, School of Natural Sciences, Newcastle University, UK. NE1 7RU.
DNA templated assembly of semiconudctor material and metals has been an area of recent interest with applications in sensors and molecular electronics.1 Recent work in the Chemical Nanoscience Laboratories has reported the inclusion of redox active nucleotides into solvated and silicon bound ssDNA.2 Hybridisatioin events were detected electrochemically and the potential for gene sensing demonstrated.
The initial studies into the incorporation of conductive polymer units into modified nucleosides for assembly at ssDNA are described here. The synthesis and crystal structure of propyn-tpt (thiophene-pyrrole-thiophene) 1, and the coupling of this monomer unit to thymidine at the C-5 position is reported. AFM studies showed that 1 was able to hybridise to strands of the poly-adenine oligomer. Prevoius work had shown that polypyrrole could be immobilised onto a DNA modified silicon electrode through electrostatic interactions between the two polymers.3 Here preliminary cyclic voltammetry studies also indicated that 1 could be polymerised when hydrogen bonded to a short strands of poly-A covalently bound to a silicon electrode. This work demonstrates that thiophene-pyrrole-thiophene modified monomers of DNA can be polymerised whilst hydrogen bonded to long strands of ssDNA. DNA shows promise as a site-directing scaffold onto which highly defined conducting nanowires can be assembled.
[1] Carrell et al, Angew. Chem. Int. Ed. 2007, 46, 6226-6236.
[2] Pike et al, Chem. Eur. J. 2005, 11, 344-353.
[3] Pike et al, Adv. Mater, 2003, 15, 254-257.
1 University of Aarhus, 2 Arizona State University
Jacob Ask Hansen, ask@chem.au.dk, University of Aarhus, Chemical department, kangelandsgade 140 building 1510, 8000 Aarhus C
Metal sulfide nanoparticles have been applied for the design of three sensor setups for the amplification and electrochemical detection of biomolecular interactions. Two sensors were developed using a novel displacement assay, one for the detection of DNA1 and one for the simultaneous detection of multiple proteins2 (see Fig 1).
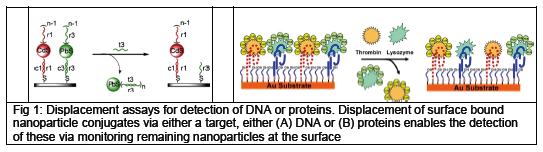
The nanoparticle conjugated reporters (DNA (A) or proteins (B)) were attached to a gold surfaces using hybridization to capture sequences (for DNA detection) or binding aptamers (for protein detection) immobilized on the gold surface via thiol-linkers. Addition of the target displaces the reporter nanoparticle conjugates from the gold surface, and thus enabling the detection of the target via stripping voltammetry by monitoring the amount of reporter nanoparticle conjugates remaining at the surface. Using this methodology we achieved sensitivities for DNA of 10amol, and for proteins 50amol, while still maintaining good selectivity.
As an extension of the above mentioned sensors we have developed a sensor based on the estrogen receptor (ER) as the biorecognition element for the detection of estrogenic compounds (agonists and antagonists).3 Binding ligands to the ER changes its conformation. This change is reliant on the type of ligand bound and gives 3 different conformations of the ER, the native state of the ER, binding of an agonist, and binding of an antagonist. Conjugating CdS nanoparticles to small peptides recognizing the different conformations of the ER enables us to design a sensor capable of detecting and differentiating ligands binding to the ER (see Fig 2).
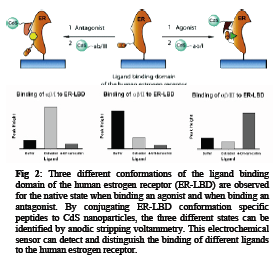
[1] J. A. Hansen, R. Mukhopadhyay, J. O. Hansen, K. V. Gothelf, J. Am. Chem. Soc., 2006, 128, 3860-3861
[2] J. A Hansen, Joseph Wang, Abdel-Nassar Kawde, Yun Xiang, Kurt V. Gothelf, Greg Collins, J. Am. Chem. Soc., 2006, 128, 2228-2229
[3] J.A. Hansen, V. V. Sumbayev, K.V. Gothelf, Nano Lett., 2007, 7, 2831-2834
University of California San Diego, Department of Bioengineering and Department of NanoEngineering, La Jolla, CA 92093-0412, mheller@bioeng.ucsd.edu
One of the grand challenges in nanotechnology is the development of fabrication technologies that will lead to cost effective nanomanufacturing processes. Over the past decade, electronic microarray devices have been used to carry out the parallel addressing and selective binding of charged biomolecules such as DNA, RNA, biotin/streptavidin, and antibodies; as well as quantum dots, metallic and polymeric nanoparticles, cells and even micron sized semiconductor devices. More recently, an electronic microarray process has been developed for the rapid and highly parallel assisted self-assembly of protein and DNA derivatized nanoparticles into multi-layer structures. This process allows 3D structures with more than forty alternating nanoparticle layers to be completed in less than one hour. Electric field assisted self-assembly represents an example of combining some of the best aspects of "top-down" and "bottom-up" technologies into viable process for the hierarchical assembly and integration of nanocomponents into 3D structures. The process is now being used for the fabrication of bio/chemsensor devices and in-vivo therapeutic/drug delivery devices; it may also prove useful for many nanoelectronic, nanophotonic, energy conversion (fuel cells, photovoltaics, batteries) and nanocomposite material applications.
In cancer research and clinical diagnostics, it is a significant challenge to directly isolate and identify rare cancer cells and potential cancer markers such as high molecular weight DNA nanoparticulates and immunocomplexes in blood, plasma and other clinically relevant samples. The advent of bio/nanotechnology has now led to numerous drug delivery approaches that involve encapsulation of drugs and imaging agents within nanovesicles and nanoparticles, which will also have to be identified and separated from blood and plasma. AC electrokinetic techniques such as dielectrophoresis (DEP) offer a particularly attractive mechanism for the separation of cells, biomarkers and drug delivery nanovesicles. Unfortunately, present DEP systems require significant dilution of the blood or plasma, thus making the technology less suitable for clinical sample preparation and diagnostic applications. Electronic DEP microarray systems have now been developed which allow separation and detection of cells, microspheres and DNA nanoparticles to be carried out under higher ionic strength conditions that more closely approach blood and plasma.
[1] Dehlinger DA, Sullivan B, Esener S, Hodko D, Swanson P and Heller MJ, "Automated Combinatorial Process for Nanofabrication of
Structures Using Bioderivatized Nanoparticles", J. Assoc. Lab Automation, October 2007.
[2] Dehlinger DA, Sullivan BD, Esener SA and Heller MJ, Electric Field Directed Assembly of Biomolecular Derivatized Nanoparticles
into Higher Order Structures, SMALL, V3, #7, pp. 1237-1244, 2007
[3] Heller, MJ, Dehlinger D, Esener SA, Sullivan B, "Electric Field Directed Fabrication of Biosensor Devices from Biomolecular
Derivatized Nanoparticles", ASME Proceedings of BioMed 2007, Next Generation Devices-38093, 2007
Fraunhofer Institute for Biomedical Engineering, Am Mühlenberg 13, 14476 Potsdam-Golm, Germany ralph.hoelzel@ibmt.fraunhofer.de
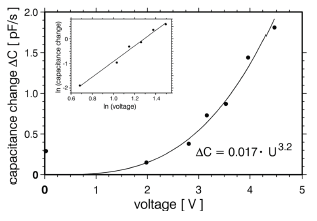
Much of the effort during the development of nanostructures is involved not so much in their preparation but in the detection and characterisation of these structures. In order to facilitate this work an electrical method has been developed that is based on the measurement of capacitance changes between microelectrodes as a result of DNA concentration changes.
This method has been known before for the investigation of micrometre-sized particles and has been introduced by us to the molecular level [1]. This approach is advantageous in so far as the molecules of interest do not need to be labelled and no optical or mechanical access is necessary as compared to microscopical methods. Above that automation is simple.
Currently the systems allows the label-free detection of 10 pg/μl DNA in samples of 2 μl volume. We have applied it to the investigation of the dielectrophoretic (DEP) response of pBlueScript dsDNA in inhomogeneous electrical AC fields. This effect allows the concentration, stretching and alignment of polarisable macromolecules and is widely used for the spatial manipulation of DNA. Usually experimental parameters are chosen rather empirically. There are only few studies aimed at an principal understanding of molecular DEP, which use fluorescence microscopy on labelled DNA. We have used the DEP field electrodes to monitor their mutual capacitance in order to study the influence of electrical as well as chemical factors on molecular DEP of pBlueScript DNA.
Capacitance measurements allow a label-free study of macromolecules. Improvement of the method's sensitivity by some orders of magnitude, possibly to a few molecules [2] appears feasible.
[1] Hölzel, R. and Bier, F. F., AIP Conf. Proc. 725, 77-83 (2004)
[2] Hölzel, R., Calander, N., Chiragwandi, Z., Willander, M., Bier, F. F., Phys. Rev. Lett. 95, 128102 (2005)
1 JBCI, Institut of Physical Chemistry, Friedrich-Schiller-University Jena, Helmholtzweg 4, D-07743 Jena, Germany; E-Mail: Juergen.Popp@uni-jena.de
2 Institut of Photonic Technology, Albert-Einstein-Straße 9, D-07745 Jena, Germany
The growing interest in DNA diagnostics is today addressed by microarrays with fluorescence detection. Unfortunately there are some disadvantages concerning multiplexing. In our approach we utilize spatially defined arrays of short oligonucleotides on a modified glass surface. For detection surface enhanced resonance Raman scattering (SERRS) is used to obtain molecularly specific spectra of the Raman active dye-labeled DNA. SER(R)S allows the construction of multilabel systems with nearly no limitations in the numbers of labels used parallel, since the limitation of non-overlapping absorption and emission spectra required for fluorescence labelling does not exsist anymore. The interaction between an analyte molecule and a roughened metallic surface in SERS leads to an enhancement of the intrinsically weak Raman signal. Thus SERS provides detection limits, which are at least as good as those of fluorescence and allows a detection of molecules down to the single molecule level.[1] As SERS active substrate nanoparticles produced by enzymatic silver deposition are utilized. They grow directly on the modified oligonucleotides and only in the spatially defined areas on the chip. The applied biotin coupling system only requires a standard modification of the oligonucleotide, which is simple, stable and cost-effective in comparison to the usually used thiol modification. Furthermore they offer several advantages for SERS detection. The nanoparticles are characterized and their potential for their use as SERS and SERRS active substrate is estimated. Therefore three different Raman active dyes are used and the potential as application for sequence specific DNA analysis is pointed out.[2]
[Left] SEM image of a silver spot after enzymatic silver deposition. The flake-like structure of the enzymatic nanoparticles is visible. [Right] Principle approach for the investigation of the enzymaticly produced silver nanoparticles as SER(R)S substrate.
[1] K. Kneipp et al., Physical Review Letters 1997, 78 (9), 1667
[2] K. Hering et al., ChemPhysChem 2008, accepted for publication
1 Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, Russia
2 Laboratory for the Physics of Nanostructures, Ecole Polytechnique Federale de Lausanne, Switzerland
3 Department of Biochemistry, Tel Aviv University, Israel
The development of a controlled molecule arranging constitutes the basis for realization of nanoscale devices such as molecular matrices, biochips and bioelectronic circuits.
We present a new method for precise positioning and alignment of DNA molecules, based on chemical patterning of surfaces induced by direct electron beam exposure. Construction of ordered arrays of aligned double-strand and triplex DNA molecules is demonstrated, and the positioning and alignment mechanisms are investigated. The direct electron beam writing on a prefunctionalized surface [1] induces nanometerscale local chemical modifications. This is combined with a macroscopic deposition of DNA from a buffer solution. The self-assembly of the DNA arrays is driven by a patterned electrostatic field generated above the surface of the substrate due to the patterned functionalized layer. The molecules select the most favorable attachment locations while self-aligning during adsorption. The flexibility of the method opens many possibilities for arranging DNA and other biomolecules in dense patterns of arbitrary configurations. [2]
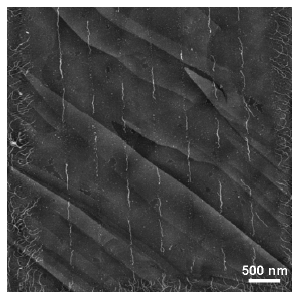
Triplex DNA molecules periodically arranged and aligned on HOPG substrate along the linear pattern exposed on NH2-terminated polymer by direct e-beam writing. Local chemical surface passivation leads to selected narrow paths where molecules can link by selecting the most favorable position during adsorption.
[1] Dmitry Klinov, Benjamin Dwir, Eli Kapon, Natalia Borovok, Tatiana Molotsky and Alexander Kotlyar, High-resolution atomic force microscopy of duplex and triplex DNA molecules, Nanotechnology, 2007, 18, 225102.
[2] Dmitry Klinov, Kirill Atlasov, Alexander Kotlyar, Benjamin Dwir, andEli Kapon, DNA Nanopositioning and Alignment by Electron Beam-Induced Surface Chemical Patterning, Nano Letters, 2007, 7, 12, 3583-3587.
1 Department of Biochemistry, George S. Wise Faculty of Life Sciences, Tel Aviv University, Ramat Aviv, 69978 Israel
2 Nanotechnology Center, Tel Aviv University, Ramat Aviv, 69978 Israel
The DNA molecule has attracted extensive interest over the past decade as a possible candidate for nanoelectronics. The self-assembling properties of DNA, its accurate synthesis, and specific interaction with proteins, guided by recognition are extremely useful for implementing self-assembly in molecular circuits. One of the main challenges with such molecules, however, is the control of their electrical conductivity. The current viewpoint is that native double stranded DNA is an insulator (poor semiconductor) rather than conductor. This prompted us to develop novel nanowires composed of either a single long (thousands of bases) self-folded G-strand [1] or four poly(G) strands clustered together by avidin. These 4G-wires comprise a large number of stacked guanine tetrads providing better conditions for π overlap compared to base-pairs of the canonical double stranded DNA. A high content of guanines, which have the lowest ionization potential among DNA bases, also makes charge migration through the DNA highly probable. These properties strongly indicate that the conductivity of G4-DNA is potentially better than that of dsDNA, making G4-DNA a valid alternative to dsDNA to develop DNA-based nano-electronics. Preliminary conductance measurements revealed very high currents flowing through single G4-DNA molecules placed between a conductive-AFM tip and a metal electrode. I-V characteristics of several G4-DNA molecules trapped between lithographically defined nanoelectrodes (100 nm distance) show significant currents as well.
We have find that incubation of the G4-DNA wires with porphyrins results in formation of a very stable complex between the DNA and the dye [2]. The complex characterized by a high ratio for porphyrin to G4 binding equal to 0.5. We have shown that the mechanism of the porphyrins binding includes intercalation of the ligand between each pair of successive G-tetrad planes in 4G-DNA. The planar porphyrin molecules are intercalated within 4G-DNA and stabilize the complex through favorable π-stacked interactions between aromatic residues, as for "classical" duplex intercalation. We showed by AFM imaging analysis that binding of the porphyrins does not increase the overall length of the wire pointing out that the average distance between the aromatic stacked planes (4G-tetrade or intercalator) in the dye-DNA complex is lower than 3.3 Å (an average distance between adjacent tetrads in the G4-DNA). The electronic coupling in the wire depends on the planes separation; reduction of the average interplane distance in the wire should strongly increase π- π electronic overlapping and lead to charge transfer in the DNA. The π-stacking between the G-tetrads and the porhyrin within the 4G-DNA-porphyrin complex might induce high conductivity of the structure and thus provide electrical properties that are suitable for molecular electronics. The conductive properties of the suggested bio-electrical molecular "nanowire" depend strongly on the nature of the intercalated organic molecule on the average distance between the stacked planes in the wire and can be varied by changing the tetrade to intercalator ratio. The ability to incorporate intercalating molecules, having different electron conducting properties into a single DNA "wire" also enables manipulation of the conductivity of the "nanowire". This will expand the applicability of the wires as elements in DNA nanochips and biosensors.
A metal-free and Zn-porphyrins are photoactive and characterized by long lasting excited triplet state. The triplet state characterized by low redox potential and is capable of abstracting an electron at electrical potentials much lower than the ground state. When complexed with 4G DNA the intercalated porphyrin molecules will carry the current through the polymer at relatively low applied electrical potentials only in the presence of light. This property is essential for development molecular electro-optical devices.
[1] Borovok N, Molotsky T, Ghabboun J, Porath D, Kotlyar A. Efficient procedure of preparation and properties
of long uniform G4-DNA nanowires. 2008 Anal Biochem. 374, 71-78
[2] Lubitz I, Borovok N, Kotlyar A. B. Interaction of monomolecular G4-DNA nanowires with TMPyP:
evidence for intercalation. 2007 Biochemistry 46 12925-12929.
1 Nanoscience Center, Department of Physics, University of Jyväskylä, P.O. Box 35, FIN-40014, Finland
2 Bell Laboratories, Alcatel-Lucent, Murray Hill, NJ 07974, USA.
3 Present address: Materials Science and Engineering Department, Boise State University, Boise, ID 83725,USA
4 Laboratory of Physics, Helsinki University of Technology, P.O. Box 5100, FIN-02015 HUT, Finland
email: ankuzyk@cc.jyu.fi
Due to its exceptional self-assembly properties, DNA could become a key player in bottom-up fabrication of nanoscale systems.[1] A striking example of a DNA self-assembly technique is "DNA origami" which involves folding long single-stranded DNA with the help of short oligonucleotides.[2] DNA molecules and DNA selfassembled structures can be uses as a template for attachment of various materials (proteins, carbon nanotubes, metal nanoparticles). In case of DNA origami each short oligo can serve as a pixel. Therefore, origami structures can be decorated with complex patterns with 6 nm resolution.
Controlled positioning of DNA molecules and origami structures on the chip is a crucial open challenge for the realization of the full potentials of DNA as template. In our previous works we have developed dielectrophoresis-based method for trapping of DNA molecules (see Fig. 1A).[3] Recently we showed that DEP can also be used to trap DNA origami structures (see Fig. 1B).[4] The method gives a high yield of single molecule/structure trapping between nanoelectrodes and controlled positioning on a chip and provides means of bridging bottom-up and top-down fabrication approaches in nanotechnology. Trapping of DNA origami structures is the first demonstration of the DEP manipulation of complex self-assembled structures.
We have also demonstrated that DNA origamis can be used for complex assembly of proteins (streptavidin) to prove the feasibility of origami as template idea (see Fig. 1C, unpublished data).
Figure 1. A) Example of DEP trapping of individual DNA molecules. B) DNA origami structure trapped using DEP. (C) DNA origami used as template for complex assembly of streptavidin. The scale bar is 100 nm.
[1] N. C. Seeman, Nature 421 (2003) 427-431.
T. H. LaBean and H. Li, Nano Today 2 (2007) 26-35.
[2] P. W. K. Rothemund, Nature 440 (2006) 297-302.
[3] S. Tuukkanen, A. Kuzyk, J. J. Toppari, V. P. Hytönen, T. Ihalainen, and P. Törmä, Appl. Phys. Lett. 87
(2005) 183102.
S. Tuukkanen, A. Kuzyk, J. J. Toppari, H. Häkkinen, V. P. Hytönen, E. Niskanen, M. Rinkiö, and P.Törmä,
Nanotechnology 18 (2007) 295204.
S. Tuukkanen, J. J. Toppari, A. Kuzyk, L. Hirviniemi, V. P. Hytonen, T. Ihalainen, and P. Törmä, Nano
Lett. 6 (2006) 1339-1343.
[4] A. Kuzyk, B. Yurke, J. J. Toppari, V. Linko, and P. Törmä. Accepted for publication in SMALL.
Institute of Photonic Technology (IPHT), Jena, A.-Einstein-Str. 9, 07745 Jena
*Fraunhofer Institut für Biomedizinische Technik (IBMT), Golm, Am Mühlenberg 13, 14476 Potsdam/Golm
The use of biomolecules like DNA for generation of defined nanowires offers a large potential for nanoelectronic applications. The resulted nanowires exhibit a highly defined biomatrix with a potential for manipulation in the nanometer-range and the ability of following metallization steps for electrical contacting. Dielectrophoresis (DEP) as basis technology was used for defined and cost-effective microintegration of these framework biomolecules [1-3]. The binding of only one single DNA molecule between microelectrode gaps was focused. For the realization of this goal, real-time detection during the entire DEP process with optical in-situ characterization of fluorescence labeled DNA-molecules was used. To identify the integration conditions, different factors and key-parameters were first tested. In a semi-empiric way, parameter complexes and matrixes were determined. Important parameters like microelectrode layout, biomolecule dimension and concentration, medium, reaction time, frequency and voltage were modified in a fixed parameter matrix for the optimized nanowire building [4]. Additionally, comparison of real measurements with a theoretical calculation of the electric field intensities was demonstrated.
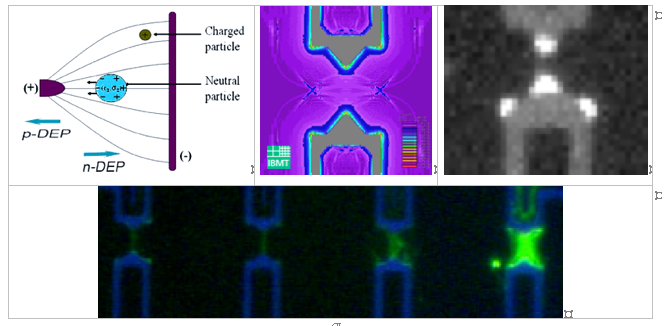
Principle of dielectrophoretis (left). Theoretical calculations of the field intensity during the DEP process (center). Field intensity spikes visualized with fluorescence labeled DNA molecules (right). Defined DNA bridges in dependence of the used voltage (bottom).
[1] Pohl, H. A. and I. Hawk (1966). "Separation of Living and Dead Cells by Dielectrophoresis." Science 152(3722): 647-649.
[2] Washizu, M. and O. Kurosawa (1990). "Electrostatic Manipulation of DNA in Microfabricated Structures." IEEE Transactions of
Industrial Applications 26(6): 1165-1172.
[3] Holzel, R., N. Gajovic-Eichelmann, et al. (2003). "Oriented and vectorial immobilization of linear M13 dsDNA between interdigitated
electrodes--towards single molecule DNA nanostructures." Biosensors and Bioelectronics 18(5-6): 555-564.
[4] Wolff, A.; Ch. Leiterer; A. Csáki; W. Fritzsche, Dielectrophoretic manipulation of DNA in microelectrode gaps for single-molecule
constructs, Frontiers in Bioscience (2008), in press
National Centre for NanoScience & Technology, No.11,Beiyitiao, Zhongguancun, Beijing 100190, China
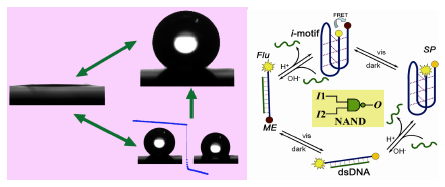
The molecular recognition properties of DNA are sufficiently well understood to enable the self-assembly of defined structures on the nanometer scale.1 DNA nanostructures have the potential to form the next generation of functional devices. An important challenge is to fabricate molecular components that act as machines.2 Based on the i-motif structure, a DNA quadruplex which is formed by DNA containing stretches of cytosine under slight acidic conditions, we have demonstrated a pH-change driven DNA nanomotor. Each working cycle of this motor could be finished in seconds and the by-product is H2O plus salt, multiple cycling of this machine has also been demonstrated.3 We have also shown the working ability of this nanomotor in the solid/liquid interface4 as well as driving a microcantilever move.5 In this report, we will report its application in fabricating smart devices such as pH trigged DNA nanocompartment6, enthalpy-driven three- State switching of superhydrophilic/ superhydrophobic surface7, as well as photo-pH dually modulated fluorescence switch and controllable assembly of gold nanoparticles.8
[1] N. C. Seeman, Nature 2003, 421, 427-431.
[2] C. Mao, W. Sun, Z. Shen, N. C. Seeman Nature 1999, 397, 144-146.
[3] D. Liu, S. Balasubramanian Angew. Chem., Int. Ed. 2003, 42(46), 5734-5736.
[4] D. Liu, A. Bruckbauer, C. Abell, S. Balasubramanian, D.-J. Kang, D. Klenerman and D. Zhou J. Am. Chem. Soc. 2006, 128,
2067-2071.
[5] W. Shu, D. Liu, M. Watari, C. K. Riener, T. Strunz, M. E. Welland, S. Balasubramanian, R. A. McKendry, J. Am. Chem. Soc. 2005,
127, 17054-17060.
[6] Y. Mao; D. Liu; S. Wang; S. Luo; W. Wang; Y. Yang; Q.Ouyang; L. Jiang Nucleic Acids Research 2007, 35, e33.
[7] S. Wang, H. Liu, D. Liu, X. Ma, X. Fang, and L. Jiang Angew. Chem., Int. Ed. 2007, 46, 3915-3917.
[8] Wenxing Wang, Huajie Liu, Dongsheng Liu, Yun Xu, Yang Yang, and Dejian Zhou Langmuier 2007, 23, 11956-11959.
Department of Biochemistry, George S. Wise Faculty of Life Sciences, Tel Aviv University, Ramat Aviv, 69978 Israel
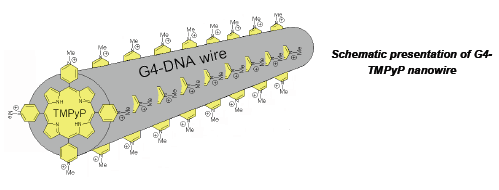
Self-assembled DNA nanostructures were suggested to have key potential in nanotechnological devices and applications. DNA nanostructure like Guanine tetrads were proposed as building blocks of molecular nanowires. The wires that we invented and used in this work comprise a large number (hundreds) of stacked guanine tetrads, providing better conditions for π-overlap compared to base pairs of the canonical double stranded DNA. High content of guanines, which have the lowest ionization potential among DNA bases, also makes charge migration through G4-wires highly probable. We have recently demonstrated that the wires are characterized by higher charge mobility as compared to double-stranded DNA. This observation makes G4-DNA a promising candidate for nanoelectronic applications. The goal of this research was to produce long conductive wires based on G4-DNA and complexes of the DNA with various intercalators. Intercalation of the aromatic molecules into the core of the DNA might increase the π-stacking among the aromatic Gtetrad planes, and as a result to improve the conductivity of the wires.
Here we present the absorption, CD and fluorescence spectroscopy data on interaction of cationic porphyrin, TMPyP (Lubitz, I., Borovok, N. and Kotlyar, A.B. 2007 Biochemistry. 46, 12925-12929) and Thiazole Orange with long monomolecular wires. The results clearly show that the molecules intercalate in-between the tetrads in the wire. The estimated ratios between the tetrad and the intecalator are 0.5 for TMPyP and 1 for Thiazole Orange respectively. TMPyP and Thiazole Orange are photoactive, their triplet states are characterized by a low redox potential and are capable of abstracting an electron at electrical potentials much lower than those of the ground state. This property of the dyes will allow current to flow through the DNA-intercalator complexes at relatively low applied electrical potentials in the presence of light. Developing of stable complexes of G4-wires with intercalators which are capable of reversibly changing their electrical conductivity upon photoirradiation is useful for application of the wires in electro-optical devices.
Department of Chemical Engineering, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong
Corresponding to: Yongli Mi, keymix@ust.hk
Figure 1: The scheme of capture and release of thrombin by sol-gel transition. (Wei B., Cheng I., Luo Q., and Mi* Y., Angew. Chem. Int. Ed. 47, 331, (2008))
In this study, a DNA-induced sol-gel transition system was combined with a protein-binding aptamer in order to capture and release proteins. Human α-thrombin was selected as the target protein and a specific thrombin-binding aptamer (GGTTGGTGTGGTTGG), which was able to form a double-stacked G-quadruplex with a high affinity to α-thrombin, was chosen for this study. The polyacrylamide hydrogel was prepared in two steps: (1) two kinds of 20mer DNA strands (G1 and G2) were grafted on the main chains of polyacrylamide; (2) polyacrylamide main chains were crosslinked into hydrogel by a 85mer DNA strand (A) with the two ends being complementary to strand G1 and strand G2, respectively. We designed the sequence of DNA strand (A) with a thrombin-binding aptamer segment, so that the thrombin can be captured by the hydrogel matrix. When we added another 85mer DNA strand, which was fully complementary to strand A, the hydrogel was dissolved and the thrombin was released.
Technische Universität Dortmund, FB Chemie, Biologisch Chemische Mikrostrukturtechnik, Otto Hahn Str. 6, D-44221 Dortmund
christof.niemeyer@tu-dortmund.de
The DNA-directed assembly of proteins offers a promising route to generate ordered multienzyme complexes with spatial control on the nanometer length scale, which are not accessible by conventional chemical crosslinking or genetic engineering.[1, 2] Here we report on the formation of an artificial bienzymatic complex, comprised of DNA-conjugated Horseradish Peroxidase [3, 4] and Glucose Oxidase. The system was characterized by gelelectrophoresis, kinetic measurements in solution as well as immobilized on surfaces and by electrochemical means. Such supramolecular structures may serve as a model system for molecular production lines, performing multistep chemical reactions. Moreover, applications in miniaturized biosensors and lab-on-a-chip devices can be foreseen.
[1] Niemeyer, C. M. (2007), Nanotoday 2, 42-52.
[2] Niemeyer, C. M., Koehler, J., Wuerdemann, C. (2002), ChemBioChem 3, 242-245.
[3] Fruk, L., Müller, J., Niemeyer, C. M. (2006), Chem. Eur. J. 12, 7448-7457.
[4] Fruk, L., Müller, J., Weber, G., Narváez, A., Domínguez, E., Niemeyer, C. M. (2007), Chem. Eur. J. 13, 5223-5231.
1 Danish National Research Foundation: Centre for DNA Nanotechnology (CDNA) at the Interdisciplinary Nanoscience Center (iNANO), University of Aarhus, DK-8000 Aarhus,Denmark.
2 Department of Molecular Biology, University of Aarhus, DK-8000 Aarhus, Denmark.
3 Department of Physics and Astronomy, University of Aarhus, DK-8000 Aarhus, Denmark.
4 Department of Chemistry, University of Aarhus, DK-8000 Aarhus, Denmark.
5 Wilhelm Johannsen Centre for Functional Genome Research, Department of Cellular and MolecularMedicine, University of Copenhagen, Denmark.
+ These authors contributed equally to this work.
We present the design and visualisation of a DNA-origami structure with flexible properties. The structure was made using our newly developed software for semi-automated design and modelling of DNA origamis. Our design, witch has the shape of a bottlenose dolphin, is an example of a nano structure with surface chiral properties and a flexible region. The dolphin was analyzed by means of high-resolution liquid AFM imaging. A high yield of DNA dolphins was observed on mica surfaces with a fraction of the dolphin nanostructures showing extensive tail flexibility of approximately 90 degrees. In addition we demonstrate a higher order assembly of two dolphins using DNA sequence specific attachment sites. The software used in the design and modelling process is available at http://www.cdna.dk/origami and freely distributed under the GNU license.
University of Copenhagen, Department of Cellular and Molecular Medicine, Blegdamsvej 3, 2200 Copenhagen, Denmark
pen@imbg.ku.dk
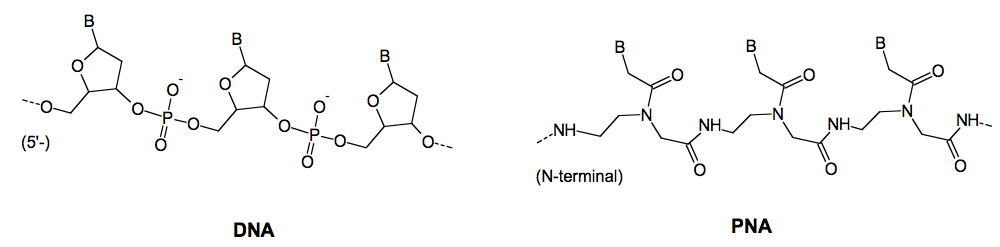
Peptide nucleic acid (PNA) is a DNA mimic based on pseudopeptide chemistry. PNA has retained the sequence recognition properties of DNA and may by virtue of these properties be key components in nanomaterials. PNA oligomers can also be designed to bind with exquisite specificity and extreme affinity to duplex DNA molecules.
This project exploits the unique DNA recognition properties of PNA to design and create systems in which DNA nanostructures self-assemble in a predetermined, sequence instructed way mediated by PNA recognition.
Homopyrimidine PNA oligomers, especially when constructed as "bis-PNAs" bind with extreme affinity and stability to sequence complementary homopurine targets in duplex DNA by a mechanism termed triplex invasion of the DNA double helix. Two PNA strands are involved both binding to the homopurine DNA strand. Thus a bis-PNA in which the two necessary PNA strands are chemically joined will effectively clamp the DNA.
An instructed DNA grid may be assembled from four DNA molecules, each containing two PNA binding sites, and the corresponding four PNA clamps.
In another approach we construct a simple DNA clover leaf composed of a circular DNA molecule with four PNA binding sites, four cognate PNA clamps (biotinylated), using streptavidin to assemble the nanostructure.
Department of Economics, Fuji University 1,
Department of Biochemistry, University of Tokyo 2,
Department of Computer Science, University of Tokyo 3
nisikawa@fuji-u.ac.jp, ohtake@biochem.s.u-tokyo.ac.jp, {fumi95, hagiya}@is.s.u-tokyo.ac.jp
The DNA walker is one of the most important applications in the field of DNA nanomachine. Typical driving force for DNA nanomachine is the so called "DNA fuel". However, some other kinds of fuel for DNA nanomachine have been developed. Takahashi et al. introduced azo-benzene intercalated DNA oligomers into DNA nanodevices which can be controlled by irradiation of ultraviolet/visible light [1]. We call this driving force photo-fuel for DNA nanomachine. We combined two more kinds of fuel with the photo-fuel[2]. They are thermo-fuel and pH-fuel. Thermo-fuel is based on the thermal control on the hybridization of DNA. On the other hand, pH-fuel is based on the structual change of the so called "i-motif" sequence, which adopts a folded form and does not hybridize with its complementary counterpart under acidic condition. These three kinds of fuel can work almost independently of one another. The present study tries to apply the method to DNA tiles.
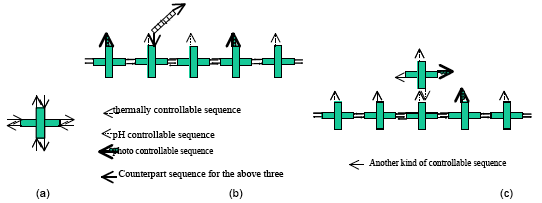
Figure 1: (a) the basic DNA cross tile. (b) the DNA trail and the simple DNA walker. (c) the DNA trail and the DNA walker made of a DNA cross tile.
Figure 1 shows the basic idea of the DNA walker and the DNA trail. A DNA cross tile has eight sticky ends as in Figure 1(a). By arranging the eight sequences, we can control the self-assembly of DNA cross tiles. We have just started to assemble DNA trails made of one kind of DNA cross tile, and we are trying to observe them by Atomic Force Microscopy (AFM) under dry condition. Currently, we have not succeeded in taking the AFM image of DNA trails. However, we got an AFM image of DNA nanotubes made of DNA cross tiles which have azo-benzene sticky ends shown in Figure 2. DNA trails are expected to be assembled by the similar protocol to that of DNA nanotubes.

Figure 2: An AFM image of DNA nanotubes made of DNA cross tiles with azo-benzene sticky ends under dry condition.
As only three kinds of fuel are available, we are working with DNA trails made of three kinds of DNA cross tiles. However, if we develop one more kind of controllable sequence, a DNA cross tile itself can be an alternative DNA walker as in Figure 1(c).
We have just started the research towards DNA walkers and DNA trails based on DNA tile assembly. We are mainly investigating DNA trails made of DNA cross tiles. Our final goal is to implement multi-fueled DNA walkers and DNA trails made of DNA tiles and to observe them with AFM directly.
[1] K. Takahashi, S. Yaegashi, H. Asanuma and M. Hagiya: Photo- and Thermoregulation of DNA Nanomachines, preliminary
proceedings of DNA11, pp.147-156(2005).
[2] A. Nishikawa, S. Yaegashi, F. Tanaka, K.Ohtake and M.Hagiya: Multi-fueled Approach to DNA Nano-robotics, Natural Computing,
to appear.
University of Bologna, Department of Biochemistry "G. Moruzzi," Via Irnerio, 48 Bologna 40126 Italy e-mail: giampaolo.zuccheri@unibo.it

collect soluble functional units at predefined locations
Complex function arises in biology from the proximity and relations amongst different functional units. Often, separate containers are employed in order to segregate specialized function within a cell, or in order to control reactions by facilitated substrates and products diffusion. Self-assembled nucleic-acids nanostructures can serve as templates for the designed assembly of different functional units that can be stably attached at locations defined with nanometer accuracy.
In this poster, a few examples are presented where the self-assembled DNA parallelogram, originally designed by Ned Seeman (Sha, Liu et al. 2000), can be used as a relatively rigid template for the assembly of different functional elements, such as oligonucleotides, fluorophores, proteins.
Depending on its structure, the functional units can be included in the assembly of the parallelogram, or added later after the assembly is completed. This yields the possibility of building nanostructures with a number of different hierarchical levels of complexity, for instance allowing recycling of the functional units without need of breaking down the more complex parallelogram structure. DNA parallelograms can also be polymerized, yielding the possibility of preparing 1D or 2D multi-functionalized templates with controlled distance between multiple functional units (Brucale, Zuccheri et al. 2006).
As in the case of fluorophores, it can be shown that the rigidity and addressability of the parallelogram tile enables a high degree of control of the relation among the functional units: the measured FRET between two organic dyes is greater than in other less flexible structures where the dyes could in principle be located at the same distance one from the other.
As these templates can bind multiple different elements, controlling their location and interaction, it is in principle possible to design functional nano-objects such as smart toxins (by assembling lytic enzymes with homing peptides or antibodies), small enzyme nanofactories (keeping multiple enzymes close in space) or smart-binders (multifunctional rigid binders that can bind and select particular conformational states of macromoleculas).
Brucale, M., G. Zuccheri, L. Rossi, A. Bazzani, G. Castellani and B. Samori (2006). "Characterization and modulation of the hierarchical
self-assembly of nanostructured DNA tiles into supramolecular polymers." Organic & Biomolecular Chemistry 4(18): 3427-
3434.
Sha, R., F. Liu, D. P. Millar and N. C. Seeman (2000). "Atomic force microscopy of parallel DNA branched junction arrays." Chem Biol
7(9): 743-51.
Chemical Nanoscience Laboratories
School of Chemistry
Newcastle University
Newcastle upon Tyne, UK
As a means of controlling the synthesis of nanomaterials DNA is increasingly considered as a material for nanotechnology. Some of these same strategies are being used to prepare nanomaterials, including nanowires, through templating reactions. However the intrinsic conductivity of DNA is not sufficient for device applications at reasonable length scales, thus precluding bare DNA as an active component in photo/electronic nanoscale devices.
Therefore we report here the synthesis of supramolecular conducting nanowires from DNA and pyrrole. Oxidation of pyrrole in DNA-containing solutions yields a material that contains cationic polypyrrole and anionic DNA. AFM imaging shows individual nanowires are continuous and around 5 nm high. They are also conformationally flexible and so can be aligned by molecular combing in a similar manner to bare DNA. This allows for the fabrication of a simple electrical device by stretching DNA/PPy strands across an electrode gap. Current-voltage measurements confirm that the nanowires are conducting, with values expected of a polypyrrole-based material. In contrast to polymerisation of pyrrole on a DNA template in solution, attempts to form similar wires by polymerisation at surface-immobilised DNA do not give a continuous coverage; instead, a beads-on-a-string appearance is observed suggesting that immobilisation inhibits the assembly.
Again using DNA strands as templates, both surface-immobilized and in solution, the growth and organization of the binary semiconductor CdS is also reported Through optimization of the reaction conditions, we have been able to control the growth and prepare quantum-confined CdS as either 1D chainlike assemblies of particles or as uniform nanowires. The latter were subsequently integrated into a simple two-terminal electrical device to demonstrate the utility of these materials as possible nanometerscale electronic components.
1- The Hebrew University of Jerusalem, Israel 2-Tel Aviv University, Israel, 3-INFM-Modena Corresponding author: porath@chem.ch.huji.ac.il
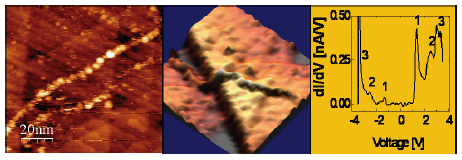
STM and STS of poly(G)-poly(C) DNA. (a) Topography image of the DNA on Gold (b). 3D presentation of the same image (c) STS on the molecule.
DNA, the most important bio-molecule, has been in the center of the scientific research for decades. In particular, DNA was considered as one of the attractive candidates for molecular electronics. DNA was, therefore, naturally chosen as one of the first investigation targets following the invention in 1982 of the scanning tunneling microscope (STM) − the first tool for direct space morphological and electrical investigation of single objects on surfaces. Attempts to resolve the energy level structure of single DNA molecules span over the last two decades, thanks to the unique ability of scanning tunneling spectroscopy (STS) to probe the local density of states of deposited objects. Nevertheless, success was hindered by extreme technical difficulties in stable deposition and reproducibility. By measuring STS on DNA at cryogenic temperature, for the first time we disclosed the energy spectrum of poly(G)-poly(C) DNA [1] and G4-DNA [2] molecules deposited on gold [3]. The tunneling current-voltage (I-V) characteristics and their derivatives (dI/dV-V) exhibit a clear gap and a peak structure around the gap. Limited fluctuations in the I-V curves are observed and statistically characterized. The character of the observed dI/dV-V peaks is assigned to orbitals originating from the different molecular components, namely the nucleobases, the backbone and the counterions, by means of ab initio Density Functional Theory calculations.
As time allows, I will show a selection of the following topics from our research: (i) Electrical measurements of relatively high current (200 nA@2 V) in short (10 nm long) DNA molecules supported by multileveled evidence. (ii) Clear polarizability of G4-DNA, a promising DNA derivative. (iii) Additional methods to measure electrical conductivity in DNA.
[1] Electronic structure of single DNA molecules resolved by transverse scanning tunneling spectroscopy", Errez Shapir, Hezy Cohen,
Arrigo Calzolari, Carlo Cavazzoni, Dmitry A. Ryndyk, Gianaurelio Cuniberti, Alexander Kotlyar, Rosa Di Felice, and Danny Porath −
Nature Materials 7, 68 (2008).
[2] "Long Monomolecular Guanine-Based Nanowires." Kotlyar, A. B., Borovok, N., Molotsky, T., Cohen, H., Shapir, E. & Porath, D.
Adv. Mater. 17, 1901 (2005).
[3] "High-resolution STM imaging of novel poly(dG)-poly(dC) DNA." Shapir, E., Cohen, H., Sapir, T., Borovok, N., Kotlyar, A. B. &
Porath, D. J. Phys. Chem. B 110, 4430 (2006).
Nanoprobes, Incorporated, 95 Horseblock Road, Unit 1, Yaphank, NY 11980, USA
* Rutgers University, Biological Sciences, 195 University Avenue, Newark, NJ 07102
Gold nanoparticles have a variety of optical, electronic, and microscopic properties that make them suitable for use as sensors. They are intensely colored, and hence may be visualized optically. Because gold possesses very high electron density, they are readily visualized and differentiated from biological materials by electron microscopy. They nucleate the deposition of metals from solution, forming the basis for the highly sensitive detection and localization of targets using autometallography. They can also interact specifically with other components of biological systems in a manner that depends upon the properties or activity of the target, thus enabling the sensing of specific chemical or biological processes with both high sensitivity and nanoscale resolution.
One of the limitations of conventional colloidal gold is that its mode of attachment to biological molecules is through non site-specific processes. By incorporating a controlled number of chemically specific reactive functional groups into the peripheral ligands of metal cluster compounds and nanoparticles, we have enabled the conjugation of these labels to biological structures at specific, unique sites where their interaction with the conjugate biomolecule is controlled at the molecular level. This property has been utilized for a variety of novel detection and sensing applications. 1.4 nm Nanogold nanoparticle labels have been conjugated to oligonucleotides at specific sites, and this allows them to be incorporated at regular sites within DNA-based nanostructured materials. When used as quenchers for molecular beacons, they provide unprecendented specificity for the detection of specific sequences and sequence mismatches in solution. Nanogold labels, linked to a substrates or cofactor of a redox-active enzyme, can accelerate the redox action of the enzyme to give enhanced sensitivity for electrochemical detection.
Gold nanoparticles may be combined with other labels to give double labeled probes. Fab' and streptavidin conjugates labeled with both Nanogold and a fluorophore have been used for correlative labeling of cellular targets for fluorescence and electron microscopy. A new class of immunoprobes labeled with Nanogold, 5 or 10 nm gold and horseradish peroxidase have been prepared. Used in conjunction with fluorescently labeled tyramides, these have demonstrated the first examples of combined fluorescent and gold labeling using a single probe with gold particles larger than Nanogold.
Methods have been developed for the covalent functionalization of larger gold particles in addition to the 1.4 nm Nanogold. Because larger 3, 5 and 10 nm gold particles have different optical and electronic properties to the smaller Nanogold cluster label, these new labels may be used to introduce new types of functionality into biological systems or nanostructured materials at specific sites, enabling the development of new sensing and detection applications.
Technische Universität, Lehrstuhl für Biologisch-chemische Mikrostrukturchemie, D-44221 Dortmund
christof.niemeyer@tu-dortmund.de
Bacterial enzymes of the cytochrome P450 family belong to the class of oxidoreductases and can catalyse a vast range of reactions.1 A very interesting feature of P450 enzymes is their ability to add a single atom of molecular oxygen to a non-activated carbon atom of a substrate, a reaction not possible by standard chemical means. Thus, they catalyze hydroxylation, epoxidation, dealkylation, or sulfoxidation reactions with a broad range of substrates. These reaction often are performed in a stereo- and/or regioselective way. Especially enzymes originating from extremophile organisms are interesting candidates for industrial applications.
CYP119, a thermostable P450 from Sulfolobus acidocaldarius, has been successfully cloned and overexpressed Echerichia coli. For the first time, it was shown that this enzyme displays peroxidase activty towards the substrate substrat AmplexRed with a range of hydroperoxides.2 Different reaction conditions, such as temperature (70°C) and pH (8.5), were optimized. These optimized reaction parameters led to 100fold increased reaction rates for conversion of the known substrate styrene. Further work concerned the synthesis and analysis of covalent CYP119-DNA-conjugates. These can be used as building blocks for nanostructured assemblies. The employment of DNA-conjugates of a thermostable enzyme, to assemble DNA-based nanostructures, is advantageous, because the assembly can also be achieved at elevated temperature without damaging the enzyme's activity. The oxidative power of CYP119 in nanostructured assemblies might be harnessed for a wide range of applications, in e.g. biosensor design.
[1] Mansuy, D. The great diversity of reactions catalyzed by cytochromes P450. Comp Biochem Physiol C Pharmacol Toxicol
Endocrinol 121, 5-14 (1998).
[2] Rabe, K. S., Kiko, K. & Niemeyer, C. M. Characterization of the peroxidase activity of CYP119, a thermostable P450 from
Sulfolobus acidocaldarius. Chembiochem 9, 420-5 (2008).
Walter Schottky Institut, Technische Universität München, Germany
We report about switchable DNA interfaces consisting of surface tethered short DNA molecules, which are driven to oscillate at high frequencies by alternating electric fields. The conformation and motion of the molecules is accurately controlled by externally applied bias, while the DNA's state is monitored in real-time by fluorescence energy transfer. We describe the preparation of switchable DNA layers and the underlying principles how oligonucleotides can be manipulated at the electrolyte/metal interface with outstanding efficiency and durability. Moreover, we demonstrate molecular dynamics measurements of the switching action by employing frequency response analyses.
The introduced methodology endows otherwise usually passive bio-interfaces with novel functionalities, and features fascinating potential for bio-sensing applications. Switchable DNA layers qualify as highly sensitive, versatile probes to detect label-free DNA as well as protein targets. In case of DNA targets, we demonstrate how binding affinities and binding kinetics can be obtained from switching measurements with single base mismatch sensitivity. Moreover, the switching behavior is shown to be a sensitive indicator for the specific recognition of IgG antibodies. Most importantly, the size of the captured protein can be inferred from the dynamics of the induced molecular motion, which allows the identification of different antibody fragments by means of their kinetic fingerprint. The switchDNA methodology represents a generic approach to simultaneously detect and size target molecules on a single analytical platform.

1: Fraunhofer Institute for Biomedical Engineering (IBMT), Department of Nanobiotechnology & Nanomedicine, 14476 Potsdam, Germany
2: University of Potsdam, Institute of Biochemistry and Biology, Potsdam, Germany
edda.reiss@ibmt.fraunhofer.de
Desoxyribonucleic acid (DNA) is regarded as a promising molecule for the bottom-up self-assembly of nanostructures.
With our approach we try to prepare long concatemeric single-stranded DNA molecules that are stretched out on a surface and consequently can be easily addressed by complementary base-pairing1. These singlestranded DNA-molecules can be regarded as the basis for the construction of a more complex DNA-scaffold.
For this purpose we have established the Phi29 DNA-polymerase mediated RCA-reaction in a flow-through system. This approach allows post-synthetic stretching of the single-stranded RCA-product as well as staining with Sybr Green II and microscopic observation without the need for disassembling the device in which the reaction is carried out. Subsequently further components like bead-tagged complementary DNA-sequences can be applied to the reaction compartment using the fluidic system without any problems and direct observation of hybridisation events is enabled via micoscopic observation.
Due to the convenience of the easy handling of our system, our approach should be a useful contribution to the world of DNA-nanoarchitecture.
[1] Reiß, E., Hölzel, R., von Nickisch-Rosenegk, M. & Bier, F. F. in DNA-BASED NANOSCALE INTEGRATION: International Symposium on DNA-Based Nanoscale Integration (ed. Fritzsche, W.) 25-30 (AIP, Jena (Germany), 2006).
Fraunhofer Institute for Biomedical Engineering, Am Mühlenberg 13, 14476 Potsdam-Golm, Germany
ralph.hoelzel@ibmt.fraunhofer.de
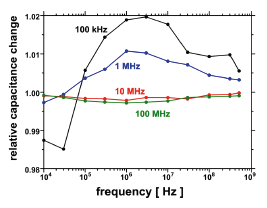
The use of DNA as a molecular brickstone is facilitated by dielectrophoretic (DEP) accumulation, stretching and alignment of the molecules. In order to quantify the molecules' DEP response and to optimise its use for nano-manipulations we have developed a detection system that exploits capacitance changes between the DEP field electrodes as a measure of local DNA concentration [1].
Interdigitated aluminium electrodes on a quartz substrate were used with 1.4 µm electrode width and 1.1 µm gap. The electrodes' mutual capacitance was measured with an impedance analyser and a test fixture. The analyser's output also served as the signal source for dielectrophoresis and was raised by an external amplifier to 3 VRMS.
pBlueScript dsDNA of 2961 bp (1.0 µm) length was prepared from a bacterial culture, linearised and dissolved in ultrapure water. Dielectrophoretic spectra were acquired automatically by repeated DEP field application for 15 s at 3 VRMS followed by 1 s to 15 s measurement at 70 to 700 mVRMS. Frequencies for DEP and capacitance measurements were varied from 10 kHz (100 kHz, resp.) to 500 MHz.
Capacitance changes were found to be maximal at the lowest measuring frequency of 100 kHz. The dielectrophoretic response of pBlueScript DNA exhibits a broad maximum between 1 MHz and 10 MHz still showing some effect at 500 MHz. Negative DEP was found below 50 kHz.
The combination of molecular dielectrophoresis and capacitance measurements allows for a rather simple experimental approach to the nanoscale spatial manipulation of macromolecules and to the monitoring of these events. The method is label-free and works well without optical or mechanical access to the probe.
[1] Hölzel, R. and Bier, F. F., AIP Conf. Proc. 725, 77-83 (2004)
1 Clinical Research Center, Department of Laboratory Medicine, Karolinska Instiutet, Novum, SE-14186 Stockholm, Sweden. 2 Universität Dortmund, Fachbereich Chemie, Biologisch-Chemische Mikrostrukturtechnik. Otto-Hahn Str. 6, D-44227 Dortmund, Germany. 3 The Judith P. Sulzberger, MD, Columbia Genome Center, Department of Physiology and Cellular Biophysics, Columbia University, New York, 10030 NY, USA. 4 Département de Chimie Moléculaire, UMR-5250, ICMG FR-2607, CNRS. Université Joseph Fourier, BP 53, 38041 Grenoble Cedex 9, France.
Correspondence should be addressed to M.G.S. (mathias.svahn@ki.se) or C.I.E.S. (edvard.smith@ki.se).
Finding the optimal combinations of ligands for tissue-specific delivery is tedious even if only a few wellestablished compounds are tested. The cargo also affects the receptor-ligand interaction, especially when it is charged like DNA. The ligand should therefore be evaluated together with its cargo.
Several viruses have been shown to interact with more than one receptor, for efficient internalization. We here present a DNA oligonucleotide-based method for inexpensive and rapid screening of biotin labeled ligands for combinatorial effects on cellular binding and uptake. The oligonucleotide complex was designed as a 44 bp double-stranded DNA oligonucleotide with one central streptavidin molecule and a second streptavidin at the terminus. The use of a highly advanced robotic platform ensured stringent processing and execution of the experiments. The oligonucleotides were fluorescently labeled and used for detection and analysis of cell-bound, internalized and intra-cellular compartmentalized constructs by an automated linescanning confocal microscope, IN Cell Analyzer 3000. All possible combinations of 22 ligands were explored in sets of 2 and tested on 6 different human cell lines in triplicates. In total, 10 000 transfections were performed on the automation platform. Cell-specific combinations of ligands were identified and their relative position on the scaffold oligonucleotide was found to be of importance.
1 Department of Life Sciences and Institute of Physics, The University of Tokyo
3-8-1 Komaba, Meguro-ku, Tokyo 153-8902, Japan
tagawa@genta.c.u-tokyo.ac.jp, suyama@dna.c.u-tokyo.ac.jp
2 School of Material Science, Japan Advanced Institute of Science and Technology
The greatest benefit of using DNA as an engineering material is its self-assembly characteristic and programmable molecular recognition ability. Especially, self-assembled DNA nanostructures are expected to be available as scaffolds for the next-generation nanofabrication. However, the problem is that the DNA scaffolds are floppy and weak as engineering materials. Therefore, it is difficult to control positioning nanocomponents with nanometer-scale precision and to manipulate with the external force.
Here, we report strong, yet flexible DNA scaffolds which are stabilized by template-directed photoligation and 5' phosphorylation [1,2]. DNA double-crossover AB-staggered (DXAB) tiles, which form the DNA scaffolds, were covalently connected by enzyme-free template-directed photoligation, which enables a specific ligation reaction in an extremely tight space and under buffer conditions where no enzymes work efficiently. Furthermore, our recent studies revealed that 5' phosphorylations contributed to the structural and thermal stabilization at the nicks. The DNA scaffolds are thus given new properties such as heat tolerance and rigidity by photoligation and 5' phosphorylation.
We additionally report programmable self-assembly method of finite-sized, fully addressable, structurallyperiodic N x M DNA arrays, which have aperiodic unique address tags on periodic DNA substrates made of only two types of DNA tiles, namely SO and SE, derived from part A and part B of the DXAB tile (Figure 1) [1,2]. The arrays were constructed in a manner similar to the way of protein synthesis, using messenger DNAs (mDNAs) that store the tile-ordering information. The substrates are completely well-ordered periodic DNA arrays due to repeats of only two kinds of tiles. The self-assembled N x M DNA arrays have the ability of the controlled positioning of matters for constructing aperiodic patterns with nanometer-scale precision.
For DNA scaffolds stabilized by photoligation, periodic substrates made up of a few kinds of building blocks are indispensable. This is because photoligation of DNA scaffolds made up of many different building blocks would bring about so complicated structural distortions that could not be removed completely. The complicated distortions may reduce the precision of nanofabrication, and at the worst break the scaffolds. The sequence design of the substrate of N x M DNA array is the same as the photoligated DNA scaffold consisting of the DXAB tiles.
The addressable photoligated N x M DNA arrays would be available as rewritable templates or scaffoldings with the ability to address DNA data-strands, nano-electronic components, and other molecules such as proteins on a nanometer scale through repeating thermal changes.
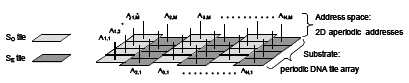
Figure 1. The design of N (row) x M (column) DNA array
[1] M. Tagawa, PhD Thesis (2007)
[2] M. Tagawa, K. Shohda, K. Fujimoto, T. Sugawara and A. Suyama, Nucleic Acids Res, 35, (21), e140 (2007)
a Graduate school of Frontier Biosciences, Osaka University, Japan.
b Institute for Photonic Technology, Germany
c Cellular Informatics Laboratory, RIKEN, Japan
d Graduate school of Frontier Biosciences, Osaka University, Japan
The molecular arrangement in nanoscale systems is of crucial importance in order to realize multi functional biosensors or molecular devices. We have developed a method for DNA-directed organization of inorganic nanoparticles. We will report the synthesis of more than 1000 bases of template DNA with a repeat unit of 100 bases by enzymatic reaction, and the fabrication of Au-nanoparticle wire. Atomic force microscope (AFM) image shows that Au-nanoparticles were ordered on specific sites by utilizing the complementarity of DNA.

Figure: AFM image (250x250 nm) of gold nanoparticles arranged along DNA template exhibiting specific binding sites
a Graduate school of Frontier Biosciences, Osaka University, Japan
b Graduate school of Frontier Biosciences, Osaka University, Japan
c Dynamic NanoMachine Project, ICORP, JST, Japan
d Graduate school of Frontier Biosciences, Osaka University, Japan
e Cellular Informatics Laboratory, RIKEN, Japan
f Department of Applied Physics, Osaka University, Japan
g Nanophotonics Laboratory, RIKEN, Japan
Nano-particles possess size-tunable optical, electrical and magnetic properties. Especially semiconductor nano-particle (Quantum dot (Q-dot)) can be used for multi-color biological imaging as well as for the construction of multi-functional biosensors and molecular devices. Arrangement of nano-particles at the molecular level is of crucial importance to realize multi-functional biosensors or molecular devices. Here we report a method for DNA-directed arrangement of Q-dot toward the fabrication of molecular devices. A template DNA more than 1,000 bases in length with a repeat unit of 100 bases was synthesized by enzymatic reaction. Alternating Q-dots alignment was fabricated by using complementarity between the template DNA and short fragments of DNA with two different sequences. Each of them was modified with two different colors of Q-dots by the avidin-biotin reaction and a reaction between an amino group and a sulfo-NHS group, respectively. These Q-dots are 5 and 8 nm in diameter, respectively. Alignment of Q-dots on the template DNA was assessed by using atomic force microscope (AFM) and transmission electron microscope (TEM). In AFM images, Q-dots were aligned at about 20 nm spacing and this extended DNA-(Qdot) complexes had heights of 3 nm on average. The observed heights of Q-dots in the AFM image were commonly lower than the actual Q-dot due to tip stress. In a preliminary experiment, alignments of two kinds of Q-dots were observed using TEM. These results provide a good starting point to the fabrication of twocolor Q-dot wire on the template DNA.
a Chemistry/Physics Dept., Univ. of New England, Biddeford, ME 04005 USA
b University of Delaware, Delaware Biotechnology Institute, Newark, DE 19711, USA
Email: jvesenka@une.edu
Guanine rich oligonucleotides (GRO) sequences common to telomeric DNA are known to create quadruplex guanine "wires", a.k.a. "G-wire" DNA. X-ray and NMR data indicate G-wires are stabilized by the incorporation of monovalent cation species between or within guanine tetramers. The telomeric forms of GRO sequences contain other nucleic acid bases common to biological sequences such as thymines (e.g. G4T2G4 found in Tetrahymena Thermophila) in which the presence of either potassium or sodium is essential for the formation of G-wires. Kmiec's work on the development of nucleic acid therapeutics involves the important discovery of guanine sequences that do not require the presence of cation stabilizers. This opens up a broad avenue of research by comparing the effect that different cationic species may have on the stabilization of G-wires and potential impact on their electronic characteristics.
We have evidence for the nature of the G-wire self-assembly using scanning probe microscopy (SPM) and circular dichroism (CD). CD spectra indicate the formation of G-wires is concentration driven (Fig. 1 − GRO in water). Scanning probe microscopy indicates that the G-wire formation involves incomplete strand pairing (Fig. 2 − GRO grown in water and directly adsorbed onto freshly cleaved mica). We examined the impact on GRO G-wire stabilization due to GRO length and cationic species with SPM. The electronic characteristics of these structures were macroscopically characterized using microelectrode masks. Electrostatic force microscopy (EFM) of the GRO G-wires attached to biased gold contacts was used to characterize their microscopic properties.
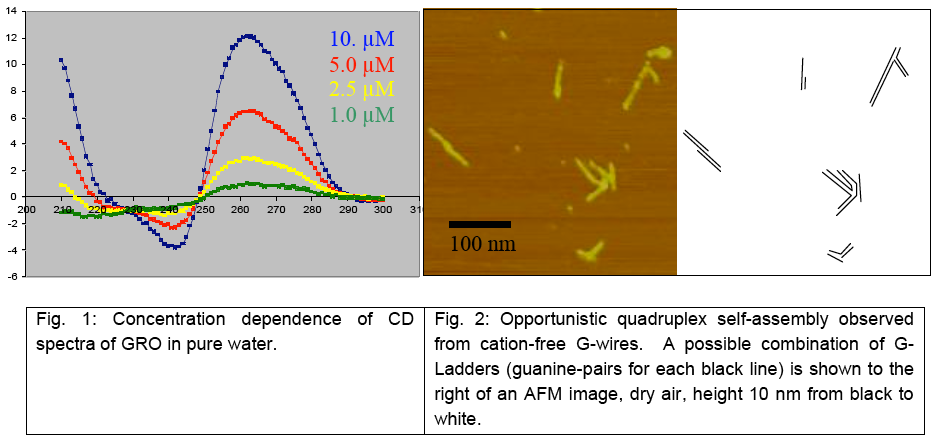
University of Bologna, Department of Biochemistry "G. Moruzzi," Via Irnerio, 48 Bologna 40126 Italy e-mail: giampaolo.zuccheri@unibo.it
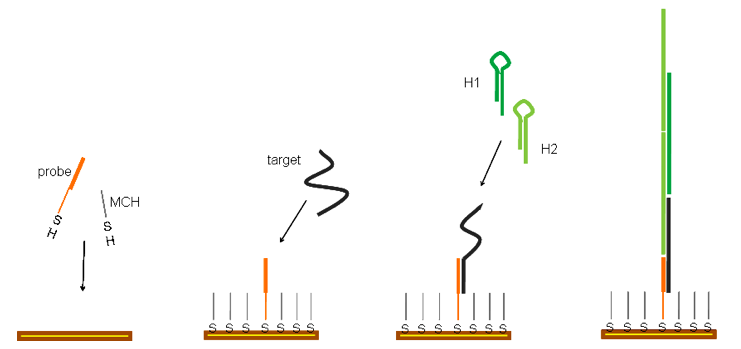
Our goal is to test simple strategies to attain high signals from DNA hybridization events. These strategies can be used towards the realization of simple, economic, portable point-of-care detection devices, to be used for diagnostics or environmental monitoring.
Hybridization Chain Reaction is an isothermal polymerization process. In this, the formation of high-molecular weight double-stranded nucleic-acid structures is triggered by the presence of a single-stranded oligonucleotide initiator (Dirks and Pierce 2004). Such polymerization needs only a limited number of oligonucleotides and no enzymes or other reactants, so it seems directly amenable of use in analytical assays.
In the preliminary experiments that we present in this poster, we tested a possible implementation of the hybridization chain reaction as a surface-bound reaction. The chain reaction is triggered by a surface DNA hybridization event, such as those involved in the recognition between a surface-bound probe oligonucleotide and a soluble analyte nucleic acid. As our current interest goes towards label-free detection strategies, we verified the success of the chain reaction both by standard techniques such as gel electrophoresis and spectrofluorimetry and also by using chemo-electronic measurements. Charged-based capacitance measurement of DNA hybridization is amenable of miniaturization for microfluidic microfabricated biosensor implementation (Guiducci, Stagni et al. 2004). Our still unoptimized results demonstrate a clear advantage in the biosensor read-out when the hybridization chain reaction is used as a pre-reading signal-enhancement step.
Dirks, R. M. and N. A. Pierce (2004). "Triggered amplification by hybridization chain reaction." Proc Natl Acad Sci U S A 101(43): 15275-
8.
Guiducci, C., C. Stagni, G. Zuccheri, A. Bogliolo, L. Benini, B. Samorì and B. Riccò (2004). "DNA detection by integrable electronics."
Biosensors and Bioelectronics 19(8): 781-787.
iPOC Research Group, Institute of Biochemistry and Biology, University of Potsdam, Germany
http://www.bio.uni-potsdam.de/nachwuchsgruppen/ipoc
Currently there is a trend from highly sophisticated analytical methods in clinical laboratories to handy and cost-effective devices for Point-of-Care Testing (POCT). Amperometric biosensors, e.g. enzyme or antibody sensors can be miniaturized, can be produced at a reasonable price and are able to measure in optically dense media. Therefore, amperometric biosensors are considered as analytical devices with a great potential for POCT.
Most of the described amperometric enzyme sensors are using oxidases as recognition elements. However, the range of detectable analytes is restricted since only a few suitable oxidases are available. In contrast to oxidases more than 400 NAD(P) dependent dehydrogenases exist. We have developed a general NAD(P)H transduction principle which can be used for the electrochemical indication of many dehydrogenase reactions. The transduction is based on a hydroxylase reaction which converts NAD(P)H to NAD(P). Thereby oxygen is consumed and an electrochemically active o-diphenol is generated which is indicated by an amperometric sensor. With this transduction principle and in combination with different analyte specific dehydrogenases it was possible to construct enzyme sensors, e.g. for the determination of glucose-6- phosphate dehydrogenase (diagnosis of hemolytic anemia) and phenylalanine (diagnosis of phenylketonuria) in blood.
By using antibodies as recognition elements the range of detectable analytes and the sensitivity can be improved further.
The hemoglobin A1c (HbA1c) value is an important clinical parameter for long term control of the glycemic status and is measured as the percentage of total hemoglobin. The clinical reference range is 5−20%, with 4−6% considered as normal. To determine the ratio of HbA1c/ total hemoglobin an amperometric sensor with immobilized anti-hemoglobin antibodies were used. After saturation of all binding sites of the capture molecule with hemoglobin of the sample the HbA1c fraction was detected via an anti-HbA1c-antibody glucose oxidase conjugate. After addition of glucose the produced hydrogen peroxide was indicated by a platinum sensor. The determined HbA1c value in the blood correlated well with the standard HPLC method.
For creatinine, which is one of the most often determined parameters in clinical diagnostics a homogenous electrochemical antibody assay was established. In the so called Size Exclusion Redox-Labeled Immunoassay (SERI) creatinine from the sample competes with a redox-labeled creatinine conjugate for the antigen binding sites of the anti-creatinine antibody. Unbound conjugate passes through a membrane (MWCO 20kD) and is indicated electrochemically at the sensor surface whereas the antibody bound conjugate is size excluded. With this wash free principle it was possible to determine creatinine in the submicromolar range.
The Hebrew University of Jerusalem, Institute of Chemistry, Jerusalem 91904, Israel
willnea@vms.huji.ac.il
Metal Nanoparticles (NPs) or semiconductor quantum dots (QDs) exhibit unique electronic, optical and catalytic properties. The synthesis of biomolecule-NP (QD) hybrid systems combine the unique physical functions of the nano-objects with the recognition/catalytic properties of the biomolecules, and this enables the use of the composite materials for sensing or nano-circuitry design. This will be exemplified with the electrical contacting of redox enzymes by means of the reconstitution of different apo-enzymes on Au NPs (1.2 nm)-cofactor units associated with electrodes.1 Alternatively, the electrical contacting of redox enzymes by means of a 3D relay-Au NP array will be described.
Different uses of biomolecule-functionalized QDs for optical biosensing will be discussed. The use of nucleic acid-functionalized CdSe/ZnS QDs for the detection of DNA and telomerase activity,2 the use of dopaminemodified QDs for the analysis of tyrosinase (a versatile marker for melanoma cancer cells),3 and the development of NAD(P)H-sensitive QDs that analyze NAD+-dependent enzymes and their substrates will be presented. The latter system was used to follow intracellular metabolism, and was applied as nano-sensor for anti-cancer cells drugs.
Biomolecule-NP conjugates provide also building blocks for the synthesis of metallic nanowires. This will be exemplified with the preparation of Au NP wires by the incorporation of intercalator (psoralen)-functionalized Au NP conjugates into a DNA template,4 and by the polymerization of Au NP-modified actin monomer units into filaments. The resulting actin-Au nanowire structures act as nanotransporters and move on surfaces upon addition of the ATP fuel.5
The nucleotide sequences in DNA encode structural and functional information into the polymers. The basesequence dictates folding motifs, catalytic properties and reactivities of the resulting DNA. These properties of DNA has been used to develop new paradigms for the amplified sensing of DNA, low-molecular-weight substrates or proteins by tailored "DNA machines".6 These biomolecular "machines" are autonomously activated upon sensing of the target analytes, and they generate a waste product that acts as the readout signal. The "DNA machines" are not only valuable analytical tools, but they enable the design of logic gates and computing systems. A DNA system that "smells-walks-computes" will be described, and the possible use of the system for therapeutics will be discussed.
The encoded information in DNA can be used to dictate the self-assembly of DNA nano-structures that yield pre-designed protein-DNA supramolecular systems. The formation of linear protein-DNA chains7 and catenated DNA chains8 will be described. Specifically, the self-assembly of hexagonic DNA wires of controlled widths will be described, and their use for activating enzyme cascades will be exemplified. Finally, DNA structures associated with electrodes act as templates for the "click-on" self-organization of solar cells. CdS NPs, photosensitizers and relays are self-assembled on DNA wires, and the resulting nanostructures reveal effective charge separation and directional photocurrent generation.
[1] (a) Y. Xiao, F. Patolsky, E. Katz, J.F. Hainfeld and I. Willner, Science, 299, 1877-1881 (2003).
(b) M. Zayats, E. Katz, R. Baron and I. Willner, J. Am. Chem. Soc., 127, 12400-12406 (2005).
[2] F. Patolsky, R. Gill, Y. Weizmann, T. Mokari, U. Banin and I. Willner, J. Am. Chem. Soc., 125, 13918-13919 (2003).
[3] R. Gill, R. Freeman, J. Xu, I. Willner, S. Winograd, I. Shweki and U. Banin, J. Am. Chem. Soc., 128, 15376-15377 (2006).
[4] F. Patolsky, Y. Weizmann, O. Lioubashevski and I. Willner, Angew. Chem. Int. Ed., 41, 2323-2327 (2002).
[5] F. Patolsky, Y. Weizmann and I. Willner, Nature Mater., 3, 692-695 (2004).
[6] (a) Y. Weizmann, M. Beissenhirtz, Z. Cheglakov, R. Nowarski, M. Kotler and I. Willner, Angew. Chem. Int. Ed., 45, 7384-7388
(2006).
(b) B. Shlyahovsky, D. Li, Y. Weizmann, R. Nowarski, M. Kotler and I. Willner, J. Am. Chem. Soc., 129, 3814-3815 (2007).
[7] Z. Cheglakov, Y. Weizmann, A.B. Braunschweig, O.I. Wilner and I. Willner, Angew. Chem. Int. Ed., 47, 126-130 (2008).
[8] Y. Weizmann, A.B. Braunschweig, O.I. Wilner, Z. Cheglakov and I. Willner, Proc. Natl. Acad. Sci., USA, in press (2008).
1Institute of Physical High Technology (IPHT) Jena, Germany;
2MPI f. Mikrostrukturphysik, Weinberg 2, Halle, Germany;
3IV. Physikal. Inst., Georg-August-Univ., Friedrich-Hund-Platz 1, Göttingen; Germany
4IHP, Im Technologiepark 25, Frankfurt (Oder), Germany
For a successful combination of Si electronics with biological applications adefined placement of biomolecules at Si surfaces is a important requirement.
One way to realize this is the using of Coulomb interaction of biomolecules with dislocations in Si. These dislocations will form charged lines in Si and will be surrounded with a space charge region being connected with an electrostatic barrier. This barrier forms an electric field reaching up to a few kV/cm.
Such a dislocation network could be formed by direct wafer bonding. In case of misorientation of the wafers, the network will be formed at the interface between them. Wafers with identical orientation would not emerge a dislocation network. An appropriate distance range, for applications in nanobiotechnology dealing with protein or DNA molecules, is from about 10 nm to a few microns. This could be adjusted by variation of the misorientation of the wafers.
We have already achieved a minimum distance between the dislocations of about 20 nm, according to transmission electron microscopy data.
With the EBIC technique the existence of a distinct electrostatic barrier/field at the interface containing the dislocation network was demonstrated. The relatively short range of the fields (smaller than one micron) necessitate the placement of the dislocation networks close to the Si surface. This could achieved with a smart cut® process whereas the wafer is thinned down from one site (smaller distances can be achieved).
In summary, we demonstrated that regular dislocation networks formed by bonding close to the Si surface might be used for self-organized placing of biomolecules. Comparable processes for biomolecule (DNA) placement will also be shown in this poster.
This project is supported by the VolkswagenStiftung.
Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA
http://people.chem.byu.edu/awoolley
We are using surface DNA assemblies in nanofabrication. Our efforts focus on making templates for the bottom-up construction of nanoelectronic circuit elements and the formation of nanocapillaries using DNA-templated nanowires. We are designing scaffolds for active circuit components using DNA origami.1 These nanoscale DNA assemblies are designed to allow them to be arranged in a controlled manner on a surface to form part of a larger device layout created by scanning probe lithography or other high-resolution patterning methods. The result will be nanoscale circuits made using a combination of top-down and bottomup methodologies. We are also using DNA to make nanometer-dimension capillaries on substrates. We recently developed the technique of DNA shadow nanolithography2 and applied this approach in creating wires, trenches and capillaries with nanometer-scale cross-sections (Figure 1). We are presently evaluating the use of DNA-templated nanowires as sacrificial materials in forming nanocapillaries. Our work demonstrates the tremendous versatility of DNA for making and arranging a variety of structures with nanometer-scale dimensions.
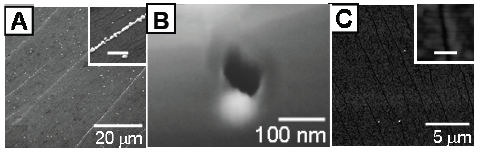
Figure 1. DNA shadow nanolithography results. (A) Electron microscopy of silver nanowires (scale bar in the inset is 2 mm). (B) Side-on electron microscopy of a nanocapillary. (C) Atomic force microscopy of nanotrenches (scale bar in the inset is 50 nm).
[1] Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 440 297-302 (2006).
[2] Becerril, H.A.; Woolley, A.T. DNA Shadow Nanolithography. Small 3, 1534-1538 (2007).
Department of Chemistry and Biochemistry & The Biodesign Institute, Arizona State University, Tempe, AZ 85287
Email: hao.yan@asu.edu
In recent years, structural DNA nanotechnology has fulfilled its promise for self-assembling both periodic and complex nanostructures1,2. One of the main objectives of this technology was to use self-assembled DNA nanostructures as designer scaffolds for directed molecular assembly. In this talk I will present our recent efforts in DNA directed self-assembly of protein3 and nanoparticle nanoarrarys4, as well as the use of selfassembling DNA nanoarrays as bioprobes5 for analytical applications and as templates for studying distance dependent biomolecular interactions.
[1] Self-assembly of symmetric finite size DNA nanoarrays, Liu, Y. Ke and H. Yan, J. Am. Chem. Soc. 127, 17140-17141
(2005).
[2] Folding DNA to create nanoscale shapes and patterns Paul W. K. Rothemund, Nature 440, 297-302 (2006).
[3] Spatially Addressable Multi-protein Nanoarrays Templated by Aptamer Tagged DNA Nanoarchitectures, Rahul
Chhabra, Jaswinder Sharma, Yonggang Ke, Yan Liu, Sherri Rinker, Stuart Lindsay, Hao Yan, J. Am. Chem. Soc. 129,
10304-10305 (2007).
[4] DNA templated self-assembly of two-dimensional and periodical gold nanoparticle arrays, J. Sharma, R. Chhabra, Y.
Liu, Y. Ke and H. Yan, Angew. Chem. Int. Ed. 45, 730-735 (2006).
[5] Self-Assembled Water-Soluble Nucleic Acid Probe Tiles for Label-Free RNA Hybridization Assays, Yonggang Ke,
Stuart Lindsay,Yung Chang, Yan Liu, Hao Yan, Science 319, 180-183 (2008).
Bioengineering Graduate Program1 and Department of Chemical Engineering2, the Hong Kong University of
Science and Technology, Clear Water Bay, Kowloon, Hong Kong SAR, China
* Corresponding author. E-mail: kehsing@ust.hk. Telephone: (852) 2358-7131. Fax: (852) 3106-4857.
In recent decades, functionalized gold nanoparticles (Au-nps) have been intensively studied as versatile reporter and carriers in developing molecular diagnostic methods, regulating intracellular gene expressions, as well as building up confined nanostructures. In particular, DNA functionalized Au-nps is one of the most attractive conjugates based on its sequence-specific molecular recognition mechanism. Among different immobilization chemistries, the direct conjugation of thiol-terminated DNA onto Au-nps via gold-sulfide bond is probably the most popular method.1 However, the charge repulsion between DNA strands and Au-nps surface and the vulnerability of unprotected Au-nps to salt-induced aggregation raise difficulties to the conjugation procedure. Generally, long incubation time (~ 2days) and careful control on ionic strength are inevitable.
In our previous study, mononucleotides, by forming an adsorption layer on particle surface, were found capable of providing a sufficient stabilization effect on Au-nps in salt solutions. This layer further demonstrated thermally dependent tunability, which became desorbed upon elevating the temperature.2 Basing on this interesting tunable stabilization methods, we will report here a new strategy to conjugate thiol- DNA to Au-nps. The overall conjugation process is significantly reduced to around 3 hours and ionic strength is no longer a concern to Au-nps in this approach. Moreover, approach will also be explored to synthesize co-conjugated Au-nps with different biomolecules, e.g. oligonucleotides and oligopeptides, which is seldom reported in the literature. Considering the important roles of nuclease and protease in disease development and molecular diagnosis, some preliminary results of a novel enzyme detection scheme will be presented utilizing the multi-functionalized Au-nps/biomolecules conjugates. We believe that the mononucleotidemediated functionalization strategy will be an attractive alternative to prepare biomolecules/Au-nps conjugates. The ability to modify Au-nps with DNAs and peptides in a controlled manner opens up new opportunities in medical diagnosis and biological sensing applications.
[1] Storhoff, J. J.; Elghanian, R.; Mucic, R. C.; Mirkin, C. A.; Letsinger, R. L., Journal Of The American Chemical Society 1998, 120,
1959-1964.
[2] Zhao, W. T.; Lee, T. M. H.; Leung, S. S. Y.; Hsing, I. M., Langmuir 2007, 23, 7143-7147.
Department of Biochemistry, George S. Wise Faculty of Life Sciences, Tel Aviv University, Ramat Aviv, 69978 Israel
DNA strands with four stretches of cytosine form an arrangement known as i-tetraplex. This structure consists of two parallel-stranded duplexes, held together by cytosine⋅cytosine+ base pairs (C⋅C+) and intercalated in an anti-parallel orientation. The physical and chemical properties of these structures are different from those of canonical double stranded DNA. In this study we synthesized novel i-tetraplex-based nanostructures composed of hundreds of C-tetrads. The method of nanostructure formation included: (1) enzymatic synthesis of long continuous dC-strands; (2) folding of the strands by pH decrease. We have demonstrated that reduction of pH to 5 results in the appearance of the characteristic bands in the CD spectrum corresponding to i-tetraplexes. We have shown that the cytosine-based nanostructures are characterized by much stronger intrinsic fluorescence compared to that of the unfolded dC-strand. The nanostructures were deposited on mica and imaged by AFM. As seen in the figure below, the molecules have a nanometric, 'bagel-shaped' arrangement with narrow diameter and height distribution. We have demonstrated that the increase of the DNA length (numbers of C-bases) results in the increase of the diameter of the nanostructures. The structures are probably originated from intramolecular folding of the Cstrand on itself. These structures are very stable and do not undergo any detectable changes during weeks at 37 °C.
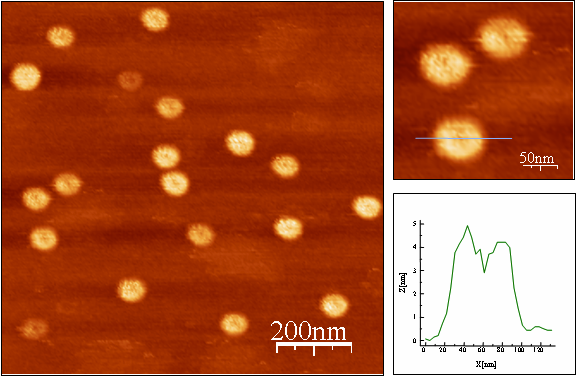
AFM imaging of C-nanostructures prepared from 1400 base dC-strand and deposited on mica. Left-bottom panel presents a cross-section of the molecule marked in the panel above.
University of Bologna, Department of Biochemistry "G. Moruzzi," Via Irnerio, 48 − Bologna 40126 Italy e-mail: giampaolo.zuccheri@unibo.it
Many reports have witnessed the potential of the atomic force microscope as a force sensor for probing the forces that hold molecules together. Accurate molecular design allowed us, amongst others, to study the unbinding forces between ligands and receptors [3] or to study protein unfolding [1, 2] and to characterize on the single-molecule level the different protein conformations present in a populations.
While there are many examples of force studies on proteins, the availability of data for pulling nucleic acids is more limited and it includes primarily optical tweezers studies. For example, processes such as the (over-)stretching of DNA or the unfolding of RNA hairpins were characterized. We are interested in extending the possibilities for the characterization of the behaviour of nucleic-acids nanostructures under force and to employ the atomic force microscope (as it can study higher forces and apply them faster). Towards this goal, we are currently trying to upgrade our force-sensing instrumentation and to design and realize an appropriate molecular design that could be helpful in the collection of force data and in its interpretation.
As previously shown by Professor Julio Fernandez, an AFM force spectroscopy apparatus can be used in a force-clamp mode, where the tensile force stretching a molecule is held constant at a pre-set value and the molecule extension is measured over time. This operational mode can yield interesting molecular information that are complementary or supplementary to those obtained with the more traditional AFM pulling mode (pulling at a constant velocity and measuring the force). In order to operate an AFM in this fashion, though, it is necessary to implement a fast force control process and to use a very fast piezoelectric actuator. In the context of a collaborative project, we built a custom-made force-clamp AFM machine based on the fastest available piezoelectric scanner. The instrument control and data acquisition is obtained through a real-time Linux application that was developed locally. The performance of this instrument has been already tested by performing protein pulling experiments on well-characterized specimens.
Force-spectroscopy experiments can be significantly simplified if the molecular bridge under tension has certain known features. First, there must be a molecular spacer, so that the interesting bond or structure is put under tension when the AFM probe is far from the substrate surface, thus avoiding non-specific forces. Secondly, it is useful to design specimens to be oligomeric in their structure, as the repetition of the same force-induced event many times in a single pulling run has distinctive advantages, as shown in the pulling of multi-modular proteins or in similarly designed systems [3]. According to this strategy, we are preparing a nucleic acid architecture designed for putting under tension a series of nanostructures and thus to study their breakdown under the application of forces (in the standard pulling mode and in the more novel force-clamp mode).
[1] Samorì, B., G. Zuccheri and P. Baschieri (2005). "Protein unfolding and refolding under force:
methodologies for nanomechanics." Chemphyschem 6(1): 29-34.
[2] Sandal, M., F. Valle, I. Tessari, S. Mammi, E. Bergantino, F. Musiani, M. Brucale, L. Bubacco and B.
Samorì (2008). "Conformational Equilibria in Monomeric α-Synuclein at the Single-Molecule Level."
PLoS Biol 6(1): e6.
[3] Valle, F., G. Zuccheri, A. Bergia, L. Ayres, A. E. Rowan, R. J. M. Nolte and B. Samorì (2008). "A polymeric molecular "Handle" for
multiple AFM-based single-molecule force measurements." Angewandte Chemie-International Edition 47(13): 2431-2434.