Home | Focus | Program | Scientific committee | Abstracts | Travel | Registration | Past Meetings
![]()
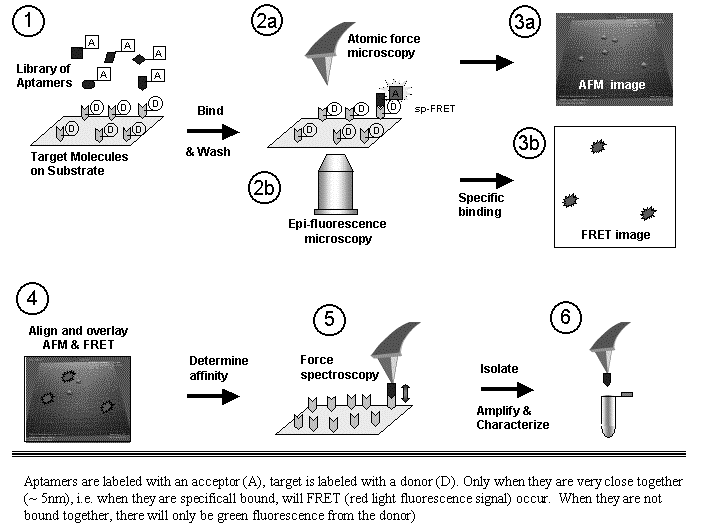
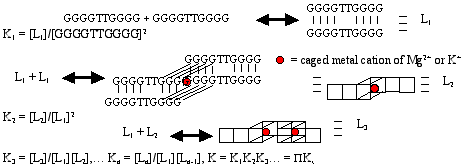
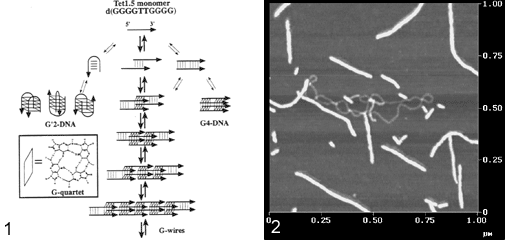
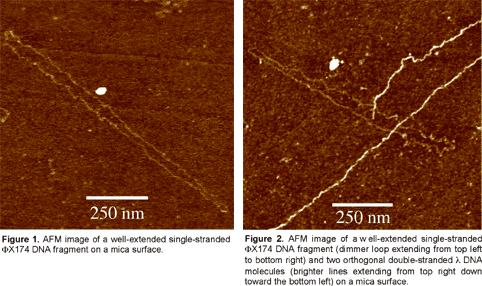
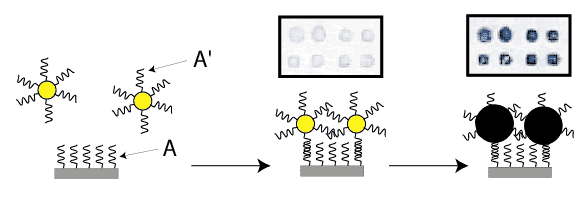
Abstracts (1 per page, 28 pages, 2.7 MB) Abstracts (4 per page, 7 pages, 1.8 MB) Nanometer addressable lateral surface structuringby use of nucleic acids Frank F. Bier, Ralph Hölzel, Nenad Gajovic-Eichelmann, Eva-Ehrentreich-Förster, Markus von Nickisch-Rosenegk, Xenia Marschan, Alexander Christmann Fraunhofer Institute for Biomedical Engineering, Dept. Molecular Bioanalytics, Bergholz-Rehbrücke, Germany For many applications manufactured surfaces with engineered structures in the nano-meter range are desirable. The properties of nucleic acids makes them a perfect material for this purpose: Nucleic acids have a regular structure with a 0.34 nm period, through its sequence it is addressable in a nanometer range. To make use of the unique features of DNA for technical applications it is necessary to link long DNA-strands in a well defined way on solid supports.Technically, DNA-oligomers are immobilised on multiple points in an ordered way. It is important to retain its functionality, the ability to hybridise to a complementary nucleic acid or to get recognised by binding proteins. Longer DNA fragments are stretched and immobilised by hybridisation between a given structure of oligo-nucleotides in the Ķm range. Here we present and compare different approaches to realise such oligo-nucleotide structures as well as demonstrate first steps to create ordered DNA-bridges between electrodes. To visualise the DNA-strand, which might be even a single one if the experimental procedure is suitable chosen, the DNA is stained by fluorochrome. An example is given in figure 1. Employed techniques include micro painting, photo-chemistry, self-organised monolayers on metal surfaces and others.Various scanning probe microscopy (SXM) methods are used to analyse these structures: Topography by AFM, scanning near field optical microscopy (SNOM) and magnetic force microscopy (MFM) are investigated with different types of labels.Beside possible applications of the DNA-modified surfaces as DNA arrays with in-gene resolution, it will provide a framework for building larger rational assemblies used e.g. in new biosensors, coupled enzyme reactions or even DNA based computing and macromolecular machines. Figure 1. DNA oriented by electrical AC field (1 MHz) between interdigitade electrodes.a. accumulation of DNA at high concentration (40 nM), b. single DNA-strands (arrows) stretched between electrodes, DNA is stained after stretching by an intercalating fluorochrome (PicoGreen). SNP Analysis by Direct Electrical Detection Jens Burmeister and Edgar Dießel Bayer AG, Central Research Department, Biophysics, D-51368 Leverkusen Single Nucleotide Polymorphisms (SNPs) are the most common type of genetic variation between individuals. SNPs promise to be particularly useful as genetic markers in the context of pharmacogenomic approaches where drug efficacy and adverse drug reactions are to be predicted on the basis of genetic tests. Thus, methods for SNP analysis are needed as a basis for personalized therapies. Direct electrical detection (DED) on planar substrates promises to be an innovative technology that enables highly sensitive analyte detection, multiplex capabilities, miniaturization (lab on a chip) and low cost. Thus, DED would be ideally suited for genetic analysis in a Point-of-Care environment. We will present our experimental work on the development of DNA assays on the DED technology platform. DNA capture probes were immobilized on silicon oxide surfaces within gaps formed by planar gold electrodes. The DNA capture probes were used for allele-specific hybridization of synthetic target molecules that were derived from gene sequences containing polymorphic sites. Following allele-specific hybridization, the duplexes were labeled with colloidal gold and subsequently enhanced by autometallographic silver deposition. Using electrical resistance measurements excellent discrimination between different alleles was achieved. Our results demonstrate the usefulness of DED technology for SNP analysis and give an example for the integration of biomolecular recognition events into silicon-semiconductor derived technology. Single-Molecule Electronics, from Carbon Nanotubes to DNA Cees Dekker Delft University of Technology, Department of Applied Physics and DIMES, Lorentzweg 1, 2628 CJ Delft, The Netherlands This talk will review some of the work on electrical transport through single molecules, as carried out in our group at Delft. I will start my talk with a brief introduction to molecular electronics and our work on carbon nanotubes and then discuss the possible use of DNA wires for molecular electronics. Carbon nanotubes are long cylindrical all-carbon molecules with unprecedented electrical and mechanical properties. I will review our recent electron-transport and STM results obtained on individual carbon nanotube molecules. Nanotubes appear to be semiconducting or metallic. The atomic structure and molecular orbitals can be studied by STM spectroscopy in nanotubes of finite length. Electrical transport has been studied through individual nanotube molecules between nanofabricated metal contacts. Nanotubes appear to be excellent coherent conductors. We have realized a variety of single-molecule devices that operate at room temperature. Biopolymers such as DNA have been proposed to act as conducting wires as well. We have carried out transport experiments on single short (30 base pairs) polyG-polyC DNA molecules between very closely spaced (10nm) metallic contacts. Nonlinear current-voltage curves indicate that DNA is a large-gap semiconductor that can be tuned to conduct carriers at very large bias voltages. At long length scales (100 nm) however, the transport currents through DNA are immeasurably small. I will show a number of experimental results from our lab and others. The prospects of using the intrinsic conductance properties of DNA for electronics are very very weak. However, DNA does allow the construction of molecular-precise circuits by self assembly. Scanning Probe Microscopes for Imaging and Manipulation of DNA Thomas Trenkler, Peter De Wolf, Irene Revenko, Craig Prater, Veeco Instruments GmbH, Mannheim, Germany; Veeco Instruments, Dourdan, France; Veeco Instruments, Santa Barbara, CA, USA; Veeco Instruments, Santa Barbara, CA, USA Atomic Force Microscopes (AFM) have been used with great success for in-situ studies of biological samples for several years now. They are gaining more and more importance as versatile tools for the manipulation and modification of biological samples in their natural environment, as well [1-4]. In this talk, we wish to review the capabilities of AFM for studies of biological samples with emphasis on imaging and manipulation of DNA. Furthermore, we wish to present some novel tool developments in the field of AFM for biological studies. First results using these novel tools will be presented, as well. After a brief introduction into Atomic Force Microscopy, we will show examples of how both Imaging and Non-imaging techniques can be used for studies of DNA. Next to gathering structural information, material properties like adhesion and viscoelasticity can be measured on the molecular level. Several examples of studies of functionalized DNA will be reviewed.In the second part of the presentation, we wish to discuss some aspects of our two novel tools PicoForce and NanoMan for studying and manipulating biological samples. Key properties of these AFM are closed loop scanning, low noise components, and integrated calculation of cantilever force constants. The PicoForce is a further development of the MultiMode AFM. It allows easier pulling exeriments where the stretching and unfolding of DNA molecules is studied. It allows as well to detect base pair interactions at the pico-Newton force range when "unzipping" DNA molecules. The NanoMan is a further development of the Dimension3100 AFM with the aim to facilitate easy manipulation of samples. We will show how DNA can be moved, bent and cut, and how DNA and other functionalized molecules can be brought together using this novel tool. In the last part of the talk, aspects of tools with multiple probes will be discussed. [1] Gimzewski, J. K. and Joachim, C. (1999). Nanoscale science of single molecules using local probes. Science. 283: 1683-1688. Handling and Manipulation of single DNA-Molecules by Kinesin-driven Microtubules S. Diez*, C. Reuther*, R. Seidel#, M. Mertig#, W. Pompe#, J. Howard* *MPI of Molecular Cell Biology and Genetics, Dresden, Germany,# Institut für Werkstoffwissenschaft, TU Dresden, Dresden, Germany Utilizing nanoscale structures of DNA as templates for metalization has proven to be a successful way to create nanowires. Prior to metalization, however, the globular DNA molecules have to be stretched and fixed to conducting contact points. For simple geometries, this has recently been achieved using a hydrodynamic flow: DNA was functionalized at one end with thiol groups that bind to gold pads on the substrate surface. The other end was stretched in the direction of the flow. Here we demonstrate a novel principle to handle and manipulate single lambda-phage DNA molecules by kinesin-driven microtubules. The technique is based on a gliding motility assay in which the substrate surface is coated with kinesin motor proteins and microtubules are propelled along the surface by the kinesin molecules in the presence of ATP. Following biotinylation of both the microtubule lattice and the ends of the DNA, a linkage can be achieved using streptavidin (Fig. 1). Using the system described above, we have demonstrated the pick-up, transport and drop- off of single DNA molecules by motile microtubules. Moreover, we observed the stretching of DNA between substrate and microtubule, as well as between two microtubules (Fig. 2). It is also possible to use the DNA to control the movement of the microtubules. To achieve this, the motor density on the substrate is lowered and the microtubules are then guided (e.g. in circles) by stretched DNA molecules that are attached to the surface with one end and that are linked to the leading (minus) end of the microtubule. Using a parallel ensemble of microtubule-based nano-handles for DNA stretching, transport and surface linkage, as well as the employment of structural selection mechanisms will allow the creation of complex and customized network structures for further metalization and nanoelectric applications. Effect of Gel Matrics on Characteriztion Fusarium oxysporum and Fusarium oxysporum f.sp vasainvectum by RAPD Analysis M. S. Khalil1, A. A. Aly1, F. Schnieder2 and K. A. Abd El-Salam1 1-Plant Pathology Research Institute, Agricultural Research Center, Egypt.2-Institute für Phytopathologie, Christian Albrechts Univ. Kiel Germany. We have used a PCR-based technique, involving the random ampilifcation of polymorphic DNA (RAPD) to access genome variability between 16 isolates from Fusarium oxysporum f.sp vasainvectum and 4 isolates Fusarium oxysporum. They were chracterized using two gel materics RAPD analysis: 8% polyacrylamide gel and 1.5% agarose gel. Good correlation was found between F oxysporum groupings obtained by polyacrylamide gel materics RAPD analysis. Resolution of DNA bands on polyacrylamide gels was superior to that on agarose gels. Silver staining was more sensitive than ethdium bromide staining. Polyacrylamide support media discriminate among F o f.sp vasainvectum and F. oxysporum isolated from cotton, which is the the most efficient procedure in terms of simplicity and rapidity. Therfore, the polyacrylamide RAPD analysis described in this paper appear to be rapid tool for the genetic characterization of a large population of Fusarium oxysporum. Key Words: Cotton, Fusrium wilt, RAPD; ethidium bromide; silver stain; agarose; polyacrylamide.. The Combination of Atomic Force Microscopy and Laser-Based Microdissection as a Tool for Molecular Biology Thalhammer, S., Hennemayer, M., Franca, L.T.C., Heckl, W.M. Institute for Crystallography – LMU, Munich, Germanywww.nano-geo.uni-muenchen.de The combination of atomic force microscopy (AFM) and laser-based microdissection provides a direct approach for the isolation of cell clusters, single cells and even cell components down to single chromosomes and chromosomal parts. Besides used as a high resolution microscope, the atomic force microscope can be used as a manipulation and isolation tool in the nanometer scale. The noncontact laser methods, laser micromanipulation and laser pressure catapulting (LMM&LPC), are used for the isolation of single cells and cell clusters from tissue sections and the isolation of genetic material. The laser precisely cuts around the selected area leaving a micron sized gap. The isolated specimen are subsequentely ejected from the slide by single laser shots onto a collection device. The isolated material can be used for further genetics analysis. In this work we present an overview of the combination of AFM and LMM&LPC for imaging, micromanipulation and microdissection of double stranded DNA and metaphase chromosomes for structural analysis of the genome organisation and molecular diagnosis. LMM and LPC were used for the isolation of: (i) single chromosomes and chromosomal fragments, (ii) detection of TT virus in single cells and tissue areas, (iii) isolation of single chroroplasts, and (iv) isolation of single tumor cells and areals from membrane mounted tissue sections. References: S. Thalhammer, R. Stark, S. Müller, J. Wienberg, W.M. Heckl (1997): The Atomic Force Microscope as a New Microdissecting Tool for the Generation of Genetic Probes. Journal of Structural Biology, 119 (2), 232-237 A construction scheme for a SET device based on self-assembly of DNA and nanoparticles W. Fritzsche, G. Maubach, D. Born, J.M. Köhler, A. Csaki Institute for Physical High Technology Jena, Biotechnical Microsystems Department Although DNA-based construction shows great promises for the realization of nanometer structures with high accuracy and reproducibility, there are numerous obstacles to overcome before it will become applicable in a technical setting. Robust processes in a highly parallel manner have to be developed to meet the requirements for a mass production at low costs. Self-assembly processes are in principle compatible with this requirements, and therefore of special interest. We proposed a scheme based on multi-level self-assembly of DNA of various length scales together with modified chip surfaces and metal nanoparticles, which is aimed at the fabrication of a single electron tunneling (SET) device on a chip surface. We will present results from the realization of different steps of this process, including different techniques for surface immobilization of longer DNA, the coupling of DNA-modified nanoparticles to long DNA, and the stretching of single DNA-molecules between two electrodes. Novel Methodology to identify single aptamer molecules Martin Guthold1, Roger Cubicciotti2, Richard Superfine3, Russell Taylor4 1Department of Physics, Wake Forest University, Winston-Salem, NC 27109, 2Nanomedica, Inc. Newark, NJ 07102, 3Department of Physics, 4Departement of Computer Science, University of North Carolina, Chapel Hill, NC 27599 Self-Assembly Driven Fabrication of Metallised DNA Nanowires and Their Electrical Properties Oliver Harnack, William E. Ford, Zoi Karipidou, Akio Yasuda, and Jurina M. Wessels Materials Science Laboratories, Advanced Technology Center StuttgartSony International (Europe) GmbH Heinrich-Hertz-Str. 1, 70327 Stuttgart, Germany We report on the development of a highly reproducible and fast method for the self-assembly driven metallisation of DNA molecules to create nanowires for applications in future nanoelectronics. The DNA metallisation was performed using water soluble, preformed gold nanoparticles that self-assemble along the DNA molecules followed by an electro-less plating step. AFM and SEM investigations (see Figure 1) were done in order to confirm the highly selective nature of our metallisation method. Measurements of the electrical conductivity of DNA nanowires across electrodes showed linear ohmic behaviour for wire networks and resistance values in the range 2 to 5 kOhm for single wires measured at 300 K. At high bias currents, clear effects of structural changes due to electromigration and overheating were obtained. A solution for the contact-less detection of conducting nanowire segments and the engineering of defects will be also part of our presentation. This work was partially funded by the European Commission through the IST-FET "Nanotechnology Information Devices" initiative, BIOAND project IST-1999-11974. Figure 1: SEM picture of metallised DNA molecules on a thermally oxidized silicon surface. DNA-directed assembly of metal nanowires Christine Keating Penn State University, University Park, PA This presentation will focus on the use of DNA to direct the assembly of metal nanowires. Nanowires are prepared by templated electrodeposition within the pores of alumina membranes. DNA has been attached to the surfaces of Au and mixed-metal nanowires several microns in length. Particle-bound DNA retains its ability to selectively and reversibly bind complementary sequences either in solution or bound to other surfaces. Thus hybridization can be used to direct the assembly of particles in solution or onto macroscopic substrates. Ultimately it may be possible to deterministically assemble nanowires into functional electronic devices in this manner. One challenge in DNA-directed nanowire assembly is preventing the formation of three-dimensional aggregates or "bundles" of particles. We are using aqueous-aqueous interfaces formed in poly(ethyleneglycol)/dextran solutions to confine nanowire assembly to two dimensions. Nanowires as large as 300 nm by 6 mm have been assembled at the interface and interrogated in situ by optical microscopy. Aqueous two-phase systems are compatible with DNA hybridization conditions (salt, temperature) and enable the DNA-driven assembly of nanowire "rafts" at the interface. Nanowire assembly and the impact of polymeric solutes on the thermodynamics of DNA-directed particle assembly will be discussed. Nanobots - just science fiction ? Günter von Kiedrowski Lehrstuhl für Organische Chemie I - Bioorganische Chemie - , Ruhr-Universität Bochum,Universitätsstrasse 150, D–44780 Bochum (Germany) The link between the fields of information based nanotechnology1 and artificial replication 2 was foreseen to lead to self-replicating molecular machinery on the nanometer scale3, called nanobots. The underlying concept was criticized from chemical and physical reasoning.4 If one however sees nanobots as threedimensionally defined noncovalent nanoscaffolds - arraying modular functions used as catalytic or binding machinery - that can be instructed to replicate by remote control, many pieces of technology needed for their implementation became recently available. Gold cluster-labelled molecules were remotely controlled by radio frequency causing local and selective inductive heating.5 Charged molecules were electrophoretically steered and manipulated on the surface of an electronic chip.6 Surface-promoted replication and exponential amplification of DNA analogues7a may find particular applications for the cloning and copying of informational nanostructures on the surface of such chips.7b Synthetic trisoligonucleotidyl junctions were reported as covalent building blocks for noncovalent DNA nanostructures and it was shown that kinetic control during noncovalent synthesis favors small and defined nanostructures instead of polymeric networks.8 Finally, functionalized DNA nanoscaffolds with stiff tensegrity such as tetrahedra were shown to self-assemble from maximally instructed sets of 3- or [3+1]-arm junctions.9 Here we demonstrate that the connectivity information in such nanoscaffolds can be copied.10 Copying of connectivity information is a purely chemical process which might be the key step for future attempts towards the replication of multifunctional nanoscaffolded machinery.References: 1. Chen, J. & Seeman, N.C. Nature 350, 631-633 (1991). On-chip solid phase DNA synthesis on semiconductor Lars H. Lie, Andrew R. Pike, Samson Patole, Benjamin R. Horrocks, Andrew Houlton, and Bernard A. Connolly University of Newcastle upon Tyne, Newcastle upon Tyne. NE1 7RU. UK. The integration of molecular materials with bulk inorganic semiconductors is important for many emerging technologies, including molecular-based electronics, nanostructured assemblies and lab-on-a-chip applications. Though DNA is often attached to silica and metal surfaces, reports on DNA-semiconductor surfaces are rare. The method we report here exploits the reactivity of hydrogen-terminated silicon surface formed by fluoride-etching. Substituted alkenes, e.g. 4,4-dimethoxytrityl-protected -undecenol, react with the hydride surface during reflux in toluene [1], and can thereafter be used as solid support for DNA synthesis. We have previously reported on-chip DNA synthesis using a commercial DNA synthesiser[2]. The fully optimised procedure used UltraMild phosphoramidites (Glen Research) and anhydrous methylamine (~2 bar) for the final deprotection step. This avoids the corrosion of the silicon that occurs under standard NH3 deprotection. A cleavable sulfonic linkage (Glen Research) was used to facilitate characterisation of the synthesised oligomer using HPLC and electrophoresis (Fig 1 and 2).On-chip synthesised oligomers were also labelled with 32P-phosphate using polynucleotide kinase and 32P-_-ATP, or hybridised to the labelled complementary strands. DNA coverage on Si(111) surface was estimated to 5.3x10-12 mol/cm2 based on electrochemical experiments using [Ru(NH3)6]3+ .This is similar to values for thiol-DNA on gold surfaces.The use of DNA as a material for nano-scale fabrication is rapidly increasing. The ability to combine this biomaterial, through the elegance of solid-phase synthesis, with semiconductor electronic materials may provide further scope for developments in nanotechnology. Literature [1] J. E. Bateman, R. D. Eagling, D. R. Worrall, B. R. Horrocks, A. Houlton, Angewandte Chemie-International Edition 1998, 37, 2683. Biomimetic fabrication of metallic nanowires and networks Michael Mertig Institut für Werkstoffwissenschaft der Technischen Universität Dresden andMax Bergmann Zentrum für Biomaterialien Dresden The construction of defined patterns of inorganic nanoparticles associated with DNA or proteins has become an inspiring field of materials science. The main goal is to combine the self-assembly capabilities of biomolecules with the quantum properties of small particles to develop new generations of devices at the nanometer scale. The peculiarity of DNA among other biological macromolecules consists in the specificity of the Watson-Crick base pairing, which allows to program their intra- and intermolecular associations and thus to build-up artificially designed supramolecular structures and networks. Moreover, DNA constitutes an ideal template for the organization of metal and semiconductor clusters into wire-like assemblies, because of its morphology, its remarkable mechanical properties, and its large variety of binding sites for different ions. We discuss two fundamental issues underlying the “bottom-up” construction of DNA- based electronic circuits: (a) the controlled integration of single DNA molecules into microelectronic contact arrays accomplished by molecular recognition between the sticky ends of DNA and specifically functionalized gold contacts, and (b) the growth of metallic Pt clusters along single DNA molecules promoted and controlled by the template itself. This method allows to fabricate ultra-thin, continuous metal cluster chains or nanowires with metallic conductivity. Moreover, first experimental evidence is obtained that the metallization kinetics is strongly influenced by the nucleotide composition of the DNA sequence. This observation suggests that selectively heterogeneous cluster nucleation and tailored template design may be combined to develop space-resolved, sequence-dependent metallization techniques in which metal structures are grown selectively on predefined portions of DNA.In addition we discuss a novel approach for the fabrication of electrical conducting networks by using motor proteins. Probing the density of surface-immobilized DNA constructs by electrical resistivity R. Möller, M. Urban, U. Klenz, H. Stürmer, W. Fritzsche Institute for Physical High Technology Jena, Biotechnical Microsystems Department Interfacing DNA constructs with surfaces is an important process for fields as chip technology and molecular nanotechnology. The efficiency of DNA construction on surfaces is a key factor in this process, but its measurement is difficult. Nanoparticle labels are one option for visualization, but the readout by scanning force microscopy (AFM) is time-consuming and so not applicable for routine use. Here we describe a setup for electrical readout of nanoparticle-labeled DNA constructs on surfaces, based on the use of chip substrates with pre-structured microelectrodes and the application of a metal enhancement procedure. Due to the ease of operation and lower equipment costs, this technique has the potential to become a routine method for measurements of the surface-density of DNA. Structure and Recognition Properties of Peptide Nucleic Acids (PNA) Peter. E. Nielsen University of Copenhagen, Institute of Medical Biochemistry & Genetics, Laboratory B, The Panum Institute, DK-2200 Copenhagen N, Denmark PNA (peptide nucleic acid) is a DNA mimic with a pseudopeptide backbone. In analogy to DNA, sequence complementary PNA oligomers form double helices, and PNA oligomers can also be designed to form other secondary structures, such as hairpins. A large variety of chemical modifications of PNA in terms of functionalised backbones and non-natural nucleobases are available. The talk will outline the chemical and structural properties pf PNA, providing examples of the chemical and structural versatility offered by the molecule that may be of use in nanotechnology development. Reference: Andrew R. Pike, Lars H. lie et al. Angew. Chem.Int. Ed..41 4, 2002. Integrating DNA With Semiconductor Materials Andrew R Pike, Lars H Lie, Samson N Patole, Lyndsey Ryder, Andrew Houlton and Benjamin R Horrocks University of Newcastle-upon-Tyne, Department of Chemistry, Bedson Building,Newcastle-upon-Tyne, NE1 7RY, UK.E-mail: a.r.pike@ncl.ac.uk Molecular electronics has become an area of considerable interest ever since the physical and economic limitations of present day microelectronic technologies have become more and more apparent. A simple and direct interconnect between silicon chips and biomolecules and the enhancement of the potential conductivity of the DNAduplex are seen to be key to future advances. Therefore a method for the automated solid-phase synthesis of DNA on a semiconductor chip with the potential for combination with photolithography to fabricate hybrid electronic-DNA devices was developed [1]. The on-chip oligonucleotide synthetic quality was comparable to standard CPG supports as confirmed by gel electrophoresis. Enzymatic manipulation of the immobilised ssDNA was possible by radiolabelling with T4 polynucleotide kinase. Spatial control, afforded by photolithography, was visualised by phosphorimaging radiolabelled dsDNA and also by electrochemical deposition of polypyrrole. The charge transfer properties of the DNA duplex were investigated by intercalation with redox active methylene blue. Additionally ferrocene modified nucleosides were incorporated into oligonucleotides to act as electronic mediators for charge transfer. Initial investigations into the effect of the redox group on the nucleobase indicated their potential for use as electronic bio-building blocks for incorporation into silicon based molecular systems. [1] A. R. Pike, L. H. Lie, R. A. Eagling, L. C. Ryder, S. N.Patole, B. A. Connolly, B. R. Horrocks and A. Houlton, Angew. Chem. Int. Ed. 2002,41, No.4, 615-617. Recognition of the DNA sequence by an inorganic crystal surface Bruno Samori*, Beatrice Sampaolese,¶ Anna Bergia,* Anita Scipioni,§ Giampaolo Zuccheri,* Maria Savino,|| Pasquale De Santis,§ *Dipartimento di Biochimica "G. Moruzzi,"Universitą degli Studi di Bologna, Via Irnerio, 48 – Bologna, Italy 40126. E-mail: samori@alma.unibo.it Differential adsorption of nucleic acid bases on graphite surface has been very recently reported (1). However, no direct evidence was so far reported of a differential adsorption of double-stranded nucleic acid molecules on a crystal surface. We obtained a clear cut evidence of a differential recognition between the surface of a crystal and the sequences that an intrinsically curved DNA molecule exposes to that surface.Curved DNA sections result planar or quasi-planar. When deposited on a flat surface, these curved DNA tracts will thus interact with one of the two faces that expose complementary sequences to the surface. The profiles assumed by these curved DNA molecules upon deposition on a flat surface can be observed by Scanning Force Microscopy (SFM). Only if the sequence orientation for each of the imaged molecules could be known precisely, then it would be possible to determine the face with which the molecules adsorb on the surface (on average) through the study of the average chain-curvature of the population of molecules. In order to remove the uncertainty on the sequence orientation in the imaged molecules, we followed the strategy of constructing palindromic DNA molecules (2). Two palindromic dimers were prepared by joining head-to-head or tail-to-tail two copies of the highly curved 211 bp segment from the Kinetoplast DNA of the Trypanosomatidae Protozoan Crithidia fasciculata, via DNA-Ligase joining of opportune restriction fragments obtained from pPK201/CAT that contains the curved segment. The molecular shapes, extracted from the SFM images were analyzed in a similar fashion as it is described in ref. (2). This made it possible to identify which face preferentially interacts with the surface. We could infer that the two dimers assume different shapes in order to expose the same sequence. The extent of this recognition effect is controlled by the degree of the curvatures. From the analysis of average local curvatures of other natural DNA palindrome dimers, we received the same evidence of the presence of this phenomenon. The recognition effect here reported is expected to be general for any curved DNA molecules.We can hypothesize that recognition processes of this kind might have been relevant in pre-cellular stages of life evolution in which inorganic surfaces might have served as templates for the self-organization of increasingly more complex bio-structures. Much effort is nowday payed to use DNA molecules for the building of self-assembling nano-structures. So far this has been done by using the self-assembling information that the sequence of these molecules contain. A higher level of information is brought into play when the same DNA molecules can also preferentially assume structures that are recognized by a crystal surface. 1. Sowerby, S. J., Cohn, C. A., Heckl, W. M. & Holm, N. G. Proc. Natl. Acad. Sci. U.S.A. 98, 820-2 (2001). DNA Nanotechnology Nadrian C. Seeman Department of Chemistry, New York University, New York, NY 10003, USA, 212-260-7905 (fax), ned.seeman@nyu.edu DNA nanotechnology uses reciprocal exchange between DNA double helices or hairpins to produce branched DNA motifs or related structures. Some of these motifs are simple branched junctions, but other motifs represent more complex strand topologies, whose structural integrity is greater. In addition to branched junctions, we have found double crossover (DX), triple crossover (TX), paranemic crossover (PX) and parallelogram motifs to be of great utility. The sequences of these unusual motifs are designed by an algorithm that minimizes sequence symmetry. We combine DNA motifs by sticky-ended cohesion, an interaction of DNA that occurs with high specificity, and that results in the formation of B-DNA; these properties make sticky-ended cohesion an outstanding means of exploiting molecular recognition, because not only is affinity guaranteed, but the local product structure is also known at the joining point. From simple branched junctions, we have constructed DNA stick-polyhedra, such as a cube and a truncated octahedron, a series of knots, and Borromean rings. We have used two DX molecules to construct a DNA nanomechanical device by linking them with a segment that can be switched between left-handed Z-DNA with right-handed B-DNA. PX DNA and a variant, JX2 DNA have been used to produce a robust sequence-dependent device (below); sequence-dependent devices can provide the diversity of structures necessary for nanorobotics. A central goal of DNA nanotechnology is the self-assembly of periodic matter. We have constructed micron-sized 2-dimensional DNA arrays from DX, TX and parallelogram motifs. We can produce specific designed patterns visible in the AFM from DX and TX molecules. We can change the patterns by changing the components, and by modification after assembly. In addition, we have generated 2D arrays from DNA parallelograms. These arrays contain cavities whose sizes can be tuned by design. We have generated DNA 6-helix bundles that cohere readily to form 1D constructs over a micron long. In addition to specific periodic self-assembly, we have performed algorithmic constructions, corresponding to XOR operations. This research has been supported by grants from the NIGMS, ONR, DARPA, NSF and USAF. DNA Nanotechnology Anett Sondermann, Claudia Holste, Robert Möller, Wolfgang Fritzsche Institute for Physical High Technology Jena, Biotechnical Microsystems Department G-wires are DNA superstructures based on the intermolecular interactions of four Guanine bases. They allow the fabrication of structures reaching the micrometer scale using only short DNA oligonucleotides, what makes them potentially interesting for molecular nanotechnology. We investigated the assembly of G-wires by SFM, using different sequences described in the literature. The assembled structures were adsorbed on mica and imaged by SFM. The influence of time and temperature of the size of the structures were studied. For one sequence we could observe 2D-aggregation beside the expected linear assemblies. Development of electronic devices based on DNA Masateru Taniguchi and Tomoji Kawai The Institute of Scientific and Industrial Research, Osaka University, Osaka 567-0047, Japan 1. IntroductionDNA is one of the important biopolymers, not only because its genetic function plays a significant role in life process, but also because its self-assembly and abilities of molecular recognition (hybridization) may give solution of problems of nano-wire and positioning at nono scale. Appropriate combination of base pairs, and modification of nanoparticles and functional molecules into DNA molecules are the promising way to control the physical properties of DNA. 2.1. Network structure(1), (2) For probing the electrical properties of Poly(dG)-Poly(dC) and Poly(dA)-Poly(dT) networks, a gold layer is evaporated by a shadow mask method to fabricate a conductive electrode. The point contact current-voltage measurements are performed by a conducting probe of AFM. The resistance is exponentially dependent on the DNA length. Poly(dG)-Poly(dC) shows better conductance than Poly(dA)-Poly(dT). 2.2. Nano-scale FET geometry(3) Current-voltage curves of Poly(dG)-Poly(dC) and Poly(dA)-Poly(dT) trapped between two Au electrodes (on Si/SiO2) separated by 20-100 nm gaps were measured at room temperature for various values of the gate voltage (Vgate). When Vgate for Poly(dA)-Poly(dT) is increased to negative values, the current is suppressed, while upon the application of a positive Vgate the current is a little enhanced. On the other hand, the current is decreased by applying a positive Vgate and increased by a negative Vgate for Poly(dG)-Poly(dC), implying that the DNA behaves as p-type semiconductor. 2.3. Chemical doped DNA thin film(4)Au electrodes with 100 nm gap were used to make electrical contacts to Poly(dG)-Poly(dC) film which were treated with the vapor of iodine at room temperature in vacuo. The value of current of I2-Poly(dG)-Poly(dC) at 1 V is enhanced up to 105 times as large as that of the non-doped film. Especially, only in case of I2-Poly(dG)-Poly(dC) film, a broad absorption peak is present around 2.6 eV, implying that charge transfer occurs between the DNA and iodine molecules. This result suggests the possibility to control the electrical properties with chemical doping. 3. Switching function with light(5)Poly(dG)-Poly(dC) has been modified by intercalation of phosphorescent molecule, acridine orange (AO). Direct measurements of electrical conductivity of AO-Poly(dG)-Poly(dC) network was performed by a conducting AFM with a gold coated tip, another gold electrode is electrically contacted on sample surface by a shadow mask deposition technique. Photoinduced electrical conductivity of AO-doped DNA network was measured by irradiating the sample with a Xe lamp (12 mW/cm2) with a cut-off filter below 400 nm. The value of electrical conductivity of AO-Poly(dG)-Poly(dC) with light irradiation is about 2-5 times as much as that without light irradiation. The result indicates that light irradiation can enhance the number of conducting carriers. It demonstrates the possibility of switching devices based on modified DNA operating at room temperature. 4. Nano patterning on Si substrate4.1. Self-organized network(2), (6), (7)Sample drops of Poly(dG)-Poly(dC) and Poly(dA)-Poly(dT) were spotted on freshly cleaved mica, and the residue solution was blew off with air. Clusters of DNA molecules were separated from each other at low DNA concentrations, while substrates were covered with a self-organized DNA network measuring more than 12 mm laterally. Furthermore, it is found that DNA molecules attach to hydrophilic SiO2 surface but not to a hydrophobic SiH surface, by controlling concentration of MgCl2. This result suggests that it is possible to fabricate micropatterning on s Si surface by using a DNA template and photolithography 4.2. Alignment of Au nanoparticles(8), (9)Gold nanoparticles have been assembled into two-dimensional complexes using a DNA network template. AFM images show that gold nanoparticles can be artificially arranged using a DNA molecular template with an average separation of 260 nm. Furthermore, the pattern of complex can be controlled by changing the concentration of the DNA solution. The results suggest that this method is effective in achieving positional control of nanoscale arrangement for a wide range of application. References Construction and Examination of “G-wire” DNA James Vesenka, Kristen Eccelston, Marcy Luhrs, Eric Henderson, & Tom Marsh University of New England, 11 Hills Beach Road, Biddeford, ME, 04005, U.S.A.E-mail: jvesenka@une.edu G-wires are four stranded DNA polymers formed by the self-assembly of simple, G-rich oligomers . G-wires are suspected to assemble through the overlap of 5’ and 3’ ends of the repeat sequence G4T2G4 (Figure 1). We have investigated the growth of G-wires using the atomic force microscope to monitor the mean length of the self-assembled molecules as a function of time (Figure 2). A system of “n” simultaneous first order linear equations is generated, requiring numerical analysis to solve for the overall constant “K”. One of the assumptions is that the addition of L1 to a G-wire chain is the rate limiting process. Keywords: G-wires, G-quartets, quadruplex DNA, scanning probe microscopy. Scanning electron micrograph of a substrate with metal-enhanced (left) and unmodified (right) 30nm gold nanoparticles. Molecular Manipulation of DNA and Its Applications Masao Washizu Department of Mechanical Engineering, Kyoto University washizu@mech.kyoto-u.ac.jp We have developed a method to stretch DNA molecule, align and immobilize onto a solid surface. The method is based on electrostatic orientation and dielectrophoresis. A pair of thin-film metal electrode is patterned onto a glass substrate, onto which DNA solution is fed, and then covered with a cover slip. The electrode is energized to 1 MV/m (100 V across 100 micron) by a high frequency (1MHz) power supply. The electrostatic field polarizes DNA molecule, and due to the interaction of the induced charge and the applied field, DNA is stretched to a straight shape. Then it is pulled into the electrode edge where the field is most intense by a phenomenon known as dielectrophoresis, and when one molecular end touches the electrode, it is anchored. The anchoring is permanent when electrochemically active metal such as aluminum is used as the electrode material. We named the process electrostatic stretch-and-positioning of DNA.Once immobilized, we can apply operations to aimed position on aimed molecule. It has been shown that desired portion of stretched DNA can be mechanically dissected, picked up, and amplified. Such a technique should open a way to "ordered" sequencing in contrast to conventional shot-gun methods in which positional information is easily lost. A microstructure has been developed in which DNA is stretched and held at both molecular termini, leaving middle part without contact to a solid surface. This allows DNA enzymes to freely interact with the immobilized DNA. The structure consists of a pair of energizing electrodes at outermost, and in between the gap are located several thin-strip electrodes having no electrical connection (floating-potential electrode, FPE). The spacing between FPEs are made slightly smaller than the length of DNA to be immobilized, and, the substrate between FPEs are etched down by a few microns. When energized, DNA molecules are stretched, and one leg is anchored onto a FPE, when the other leg reaches the adjacent FPE edge, and also anchored. DNA is held at both molecular ends bridging over the FPEs, and because the substrate between them are etched down, middle part is held without contact to the solid surface. Using FPE system, molecular surgery of DNA is demonstrated, where DNA cutting enzyme is immobilized on a microparticle, which is laser-manipulated and made into contact with stretched DNA, so that the enzymatic reaction occurs at the contact point. When DNase I is used, instantaneous dissection of DNA occurred, and when a restriction enzyme is used, dissection in agreement with the restriction map is observed. The structure also enables the observation of DNA-protein interaction on real-time, single-molecule level. With the use of fluorescence-labeled RNA polymerase, the motion of the enzyme along DNA strand as it synthesizes RNA is visualized. DNA-mediated Self-Assembly of Nanodevices Keith Williams, Cees Dekker Delft University of Technology, Department of Applied Physics and DIMES, Lorentzweg 1, 2628 CJ Delft, The Netherlands Construction of rudimentary logic circuits based on single-walled,carbon nanotube field-effect transistors (NTFETs) was recentlydemonstrated at IBM and at Delft. Our focus subsequently shifted toself-assembly strategies, which provide a means for more efficientassembly of device structures with minimal external manipulation.Clearly, an effective strategy should facilitate low-error, parallel,autonomous assembly of well-ordered structures; for this reason, we have begun work to incorporate DNA as a means for assembling nanodevices from inorganic components, such as nanotubes and fullerenes. To the "programmed" assembly schemes we envision, DNA contributes unsurpassed molecular recognition, convenient (de)hybridization conditions andsolution chemistry, and replicability. In this presentation I willdiscuss current efforts at Delft to develop a DNA-mediated assemblystrategy for nanotubes and fullerenes, and I will present prospects foralgorithmic assembly and replication. DNA Alignment, Characterization and Nanofabrication on Surfaces Adam T. Woolley, S. Darbi Hughes, Christopher F. Monson, Allison R. Nelson and Huijun Xin Department of Chemistry and Biochemistry, Brigham Young University, Provo, UT, USA.http://chemwww.byu.edu/atwoolley.php We are developing tools for surface manipulation and analysis of nucleic acid molecules for use in nanofabrication and DNA sequencing. This effort involves characterization and optimization of conditions for controlled alignment of DNA fragments on Si, mica and other substrates; and utilizing surface bound nucleic acid molecules as templates for construction of nanowires. This work builds upon our earlier results in devising methods for reproducible alignment of well-extended single-stranded and double-stranded DNA fragments on surfaces (see Figures 1 and 2).1 We have carried out a thorough study of the deposition and alignment of both single-stranded and double-stranded nucleic acids on surfaces to optimize our techniques. We are also performing single-molecule experiments to understand the underlying biophysical mechanisms that lead to directional alignment of DNA on surfaces under different conditions. Furthermore, we are using aligned, surface deposited nucleic acid molecules as templates for construction of nanowires. One approach involves photochemical and electrochemical reduction of metal ions associated with surface bound DNA to build up conductive nanostructures. Techniques for nucleic acid templated construction of carbon nanotube nanowires on surfaces are also being pursued. These results show that DNA holds great promise for fabrication of materials with nanometer dimensions, and that biotemplated nanolithography should become an important component of the nanotechnology toolbox. Studies for an optical detection of DNA constructs based on nanoparticles and silver enhancement G.-J. Zhang, R. Möller, A. Csaki, W. Fritzsche Institute for Physical High Technology Jena, Biotechnical Microsystems Department Nanoparticle-labeling was recently introduced for probing immobilized DNA by scanning force microscopy or optical detection. The optical detection has the potential of high parallelization in combination with a miniaturization, thereby enabling a high sample throughput. However, a quantification of the optical signal and a correlation of this signal with the surface density of bound nanoparticles is needed. We will present results from an investigation of different parameters influencing the optical signal amplitude and the signal-to-background ratio, using microstructured or spotted substrates. Image processing is applied in order to achieve an evaluation of the different approaches, especially regarding homogeneity and reproducibility. A Direct Measure of the Ergodicity of the Small-Scale Dynamics of DNA Giampaolo Zuccheri1, Anna Bergia1, Claudio Anselmi2, Anita Scipioni2, Pasquale De Santis2, Bruno Samori1 1Department of Biochemistry, University of Bologna, Via Irnerio, 48 – 40126 Bologna, Italy. E-mail: giampa@alma.unibo.it2Department of Chemistry, University of Roma, La Sapienza, Piazzale Aldo Moro, 5 – Roma, Italy. The slow dynamics of two differently linearized pBR322 molecules laid on a surface was monitored by time-resolved scanning force microscopy. The large-scale dynamics of the DNA molecules appears to be far from the equilibrium and retains memory of the starting structure for hours in the conditions of adsorption. On the contrary, the local dynamics well represents the equilibrium fluctuations, according to the principle of microscopic reversibility.The surface adsorption conditions of double-stranded DNA molecules on freshly cleaved muscovite mica have been optimized so that the scanning force microscopy (SFM) observations of completely hydrated DNA molecules is possible:[1] a solution of DNA molecules is spread on the surface and the molecules are observed at high resolution without the need of ever dehydrating them. In this quasi-physiologic environment, DNA molecules can be followed while they move on the surface as a consequence of the thermal agitation of the solution.Due to the strength of adsorption, the DNA molecules do not change their global shape significantly during the time of the experiments. On the other hand, the local dynamics of the chain (that can be measured through the study of the local chain curvature[2]) is significantly less hindered.The quantitative evaluation of the local chain dynamics has been performed on the basis of the ratio between the average of the modulus of the local chain angular curvature and the standard deviation of the same parameter. From theoretical considerations, such ratio SD(|Cn|)/<|Cn|> should be constant and equal to (2/(p-2))1/2. Our experimental calculations, based on the time-lapse SFM observation of 4 kbp-long DNA molecules, shows that the thermal chain dynamics over the scale of a few DNA helix turns (10–20 nm) can be considered at thermodynamic equilibrium, even though the conformation and the dynamics of the entire DNA molecule is far from equilibrium in the time-scale of the experiment. From a biological point of view, our experiment can be thought of as a measure of the condition of long DNA molecules inside the cells, where they are simultaneously involved in many interactions with other molecules and complex structures that hinder their dynamics. Our data show that each molecule that interacts with DNA on the nanometer scale can be thought of as interacting with a freely fluctuating section of the chain, even though the molecule is globally hindered. [1] Zuccheri G. et al Conformational fluctuations of supercoiled DNA molecules observed inreal time with a scanning force microscope Appl. Phys. A 66: S585-S589 (1998).![]() Download abstracts as PDF-files:
Download abstracts as PDF-files:
[2] Hansma, H. G. (1996). Atomic force microscopy of biomolecules. J. Vac. Sci. Technol. B. 14(2): 1390-1394.
[3] Bustamante, C. and Keller, D. (1995). Scanning force microscopy in biology. Physics Today. (December): 32-38.
[4] Bai, C., Zhang, P., Fang, Y., Cao, E. and Wang, C. (1997). Studies of biomaterials using atomic force microscopy. J. Korean Physical Society (Proc. Suppl.). 31: S47-S50.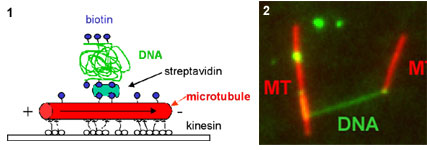
L. Schermelleh, S. Thalhammer, T. Cremer, H. Pösl, W.M. Heckl, K. Schütze and M. Cremer (1999): Laser microdissection and laser pressure catapulting as an approach for the generation of chromosome specific paint probes. BioTechniques International, 27, 362-367
P. Gobbi, S. Thalhammer, M. Falconi, R. Stark, W.M. Heckl, G. Mazzotti (2000): Correlative high resolution morphological analysis of the three-dimensional organization of human metaphasechromosomes. Scanning, 22, 273-281
S. Thalhammer, A. Kölzer, G. Frösner, W.M. Heckl (2002): Isolation of tissue sections using laser pressure catapulting for the detection of virus particles. Journal of Virology, submitted
S. Thalhammer, M. Hennemeyer, C. Meyer, H. Lorenz, G. Teti, J. Geigl, G. Mazzotti, W.M. Heckl (2002): New advances in atomic force nanomanipulation (M-AFM) – physical and biochemical parameters. Biophysical Journal, submitted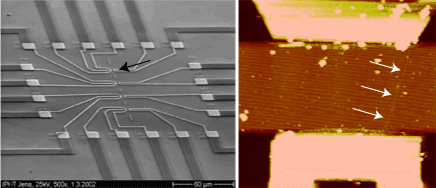

2. von Kiedrowski, G. Angew. Chem. Intl. Ed. Engl. 25, 932-935 (1986).
3. a. Drexler, K.E. Engines of creation: The coming era of nanotechnology. Anchor press, Doubleday, New York 1986; b. Winkless, N. & Browning, E. Robots on your doorstep. Robotics press, Portland 1978.
4. Smalley, R.E. Scientific American 285, 76-77 (2001).
5. Hamad-Schifferli, K. et al. Nature 415, 152-155 (2002).
6. Kricka, L.J. Nature Biotechn. 16, 513-514 (1998).
7. a. Luther, A., Brandsch, R., & von Kiedrowski, G. Nature 396, 245-248 (1998); b. von Kiedrowski, G. et al. WO ????, DE 198 54 946 C2, DE 198 48 403 A1.
8. Scheffler, M. et al. Angew. Chem. Int. Ed. Engl. 38, 3312-3315 (1999).
9. Dorenbeck, A. et al., submitted on march 12, 2002.
10. Eckardt, L. et al., submitted on march 12, 2002.
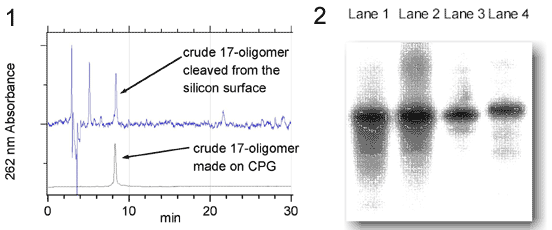
Figure 2 Polyacrylamide gel electrophoresis of oligonucleotides prepared at silicon surfaces (Lane 1 and 2)and CPG (Lane 3 and 4).
[2] A. R. Pike, L. H. Lie, R. A. Eagling, L. C. Ryder, S. N. Patole, B. A. Connolly, B. R. Horrocks, A. Houlton, Angewandte Chemie-International Edition 2002, 41, 615.
Wittung, P., Nielsen, P.E., Buchardt, O., Egholm, M. and Nordén, B.: DNA-like double helix formed by peptide nucleic acid. Nature (1994) 368, 561-563.
Rasmussen, H., Kastrup, J.S., Nielsen, J.N., Nielsen, J.M. and Nielsen, P.E.: Crystal structure of a peptide nucleic acid (PNA) duplex at 1.7 Ā resolution. Nature Structural Biology (1996) 2, 98-101.
Kozlov IA, Orgel LE, Nielsen PE: Remote enantioselection transmitted by an achiral peptide nucleic acid backbone. Angew Chem , Int Ed 2000, 39: 4292-4295.
Ganesh KN, Nielsen PE: Peptide nucleic acids: analogs and derivatives. Curr Org Chem 2000, 4: 931-943.
Kushon SA, Jordan JP, Seifert J, Nielsen H, Nielsen PE, Armitage BA: The effect of secondary structure on the thermodynamics and kinetics of PNA hybridization to DNA hairpins. J Am Chem Soc 2001, 123: 10805-10813.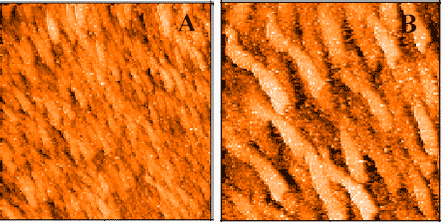
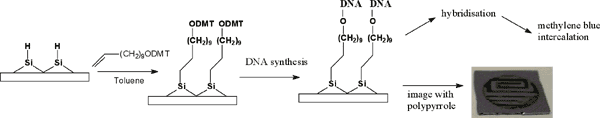
¶Centro di Studio per gli acidi nucleici del C.N.R., Piazzale Aldo Moro 5 – Roma, Italy 00185
§Dipartimento di Chimica Universitą di Roma "La Sapienza" – Roma, Italy 00185. E-mail pasquale.desantis@uniroma1.it
|| Dipartimento di Genetica e Biologia Molecolare, Piazzale Aldo Moro 5 – Roma, Italy 00185.
2. Giampaolo Zuccheri, Anita Scipioni, Giuseppe Gargiulo, Valeria Cavaliere, Pasquale De Santis, Bruno Samori' Proc. Natl. Acad. Sci. (USA, 98, 3074-3079 (2001)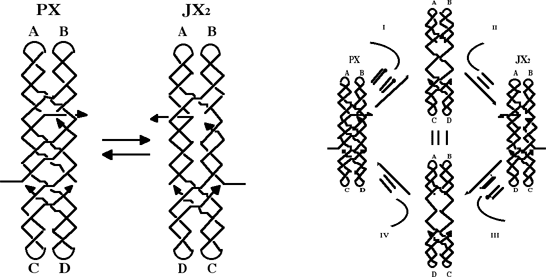
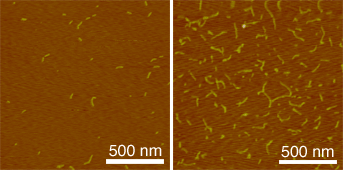
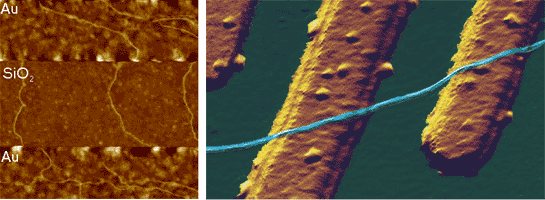
(1) L. T. Cai, et al., Appl. Phys. Lett., 77, 3105 (2000)
(2) T. Kanno, et al., Jpn. Appl. Phys., 39, L269 (2000).
(3) K. H. Yoo, et al., Phys. Rev. Lett., 87, 198102 (2001)
(4) M. Taniguchi, in preparation.
(5) J. Gu, et al., Appl. Phys. Lett., 80, 688 (2002)
(6) H. Tabata and T. Kawai, Kobunshi, 50, 251 (2001).
(7) S. Tanaka, et al., Jpn. J. Appl. Phys., 40, 4217 (2001)
(8) Y. Maeda, et al., J. Vac. Sci. Technol. B 17, 494 (1999)
(9) Y. Maeda, et al., Appl. Phys. Lett., 79, 1181 (2001).
[2] Zuccheri, G. et al. Mapping the intrinsic curvature and flexibility along the DNA chain. Proc. Natl. Acad. Sci. USA 98, 3074-9. (2001).